The pyruvate dehydrogenase complex (PDC) is one of the mitochondrial multienzyme complexes and is composed of three different enzymes:
- pyruvate dehydrogenase, or E1 (EC 1.2.4.1);
- dihydrolipoyl transacetylase, also called dihydrolipoamide acetyltransferase or E2 (EC 2.3.1.12);
- dihydrolipoyl dehydrogenase, also called dihydrolipoamide dehydrogenase or E3 (EC 1.8.1.4).
Each of these enzymes is present in multiple copies whose number, and therefore the size of the complex itself, varies from species to species, with molecular masses ranging from 4×106 to 1×107 daltons.[1]
The multienzyme complex also contains additional subunits:
- five different coenzymes;
- in plants, fungi, and, among animals, birds and mammals, two enzymes with regulatory properties: a Mg2+-dependent pyruvate dehydrogenase kinase (EC 2.7.1.99) and a Ca2+-activated pyruvate dehydrogenase phosphatase(EC 3.1.3.43);
- in eukaryotes, a binding protein called E3BP.[2]
The pyruvate dehydrogenase complex catalyzes, through five sequential reactions, the oxidative decarboxylation of pyruvate, the conjugate base of pyruvic acid, to form a carbon dioxide molecule (CO2) and the acetyl group of acetyl-coenzyme A (acetyl-CoA), with the release of two electrons carried by NAD+.[3]
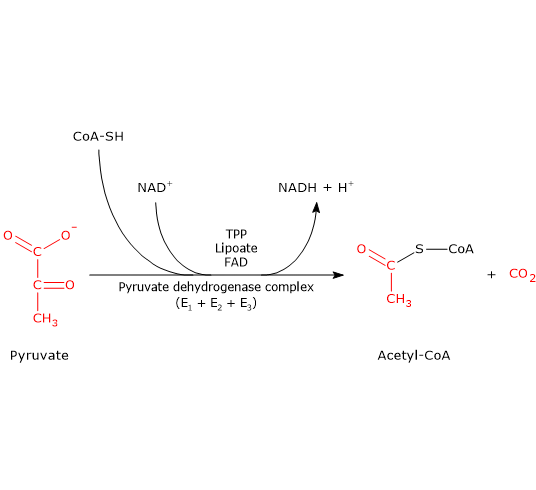
The overall reaction is essentially irreversible, with a ΔG°’ of −8.0 kcal/mol (−33.4 kJ/mol), and requires the participation of the three enzymes, whose activities are sequentially coordinated.[4] During the reactions, the intermediate products remain enzyme-bound, and at the end of the sequence the multienzyme complex is ready for the next catalytic cycle.[5]
Note: the pyruvate dehydrogenase complex catalyzes the same reactions through similar mechanisms in all organisms.[6]
Contents
- Coenzymes of the pyruvate dehydrogenase complex
- Cellular location of the pyruvate dehydrogenase complex
- Functions of the pyruvate dehydrogenase complex
- Mitochondrial pyruvate transport
- Structure of the pyruvate dehydrogenase complex
- Structure of pyruvate dehydrogenase
- Structure of dihydrolipoyl transacetylase
- Structure of dihydrolipoyl dehydrogenase
- Reaction of pyruvate dehydrogenase
- Reaction of dihydrolipoyl transacetylase
- Reaction of dihydrolipoyl dehydrogenase
- Regulation of the pyruvate dehydrogenase complex
- References
Coenzymes of the pyruvate dehydrogenase complex
Five coenzymes are involved in the reactions catalyzed by the pyruvate dehydrogenase complex. They are thiamine pyrophosphate (TPP), flavin adenine dinucleotide (FAD), coenzyme A (CoA), nicotinamide adenine dinucleotide (NAD), and lipoic acid.[1]
Thiamine pyrophosphate
Thiamine pyrophosphate is the active form of thiamine, or vitamin B1. TPP is the coenzyme of pyruvate dehydrogenase, to which it is tightly bound through noncovalent interactions. It is involved in the transfer of hydroxyethyl, or “activated aldehyde,” groups.[2]
Flavin adenine dinucleotide
Flavin adenine dinucleotide is one of the active forms of riboflavin, or vitamin B2; the other is flavin mononucleotide (FMN).
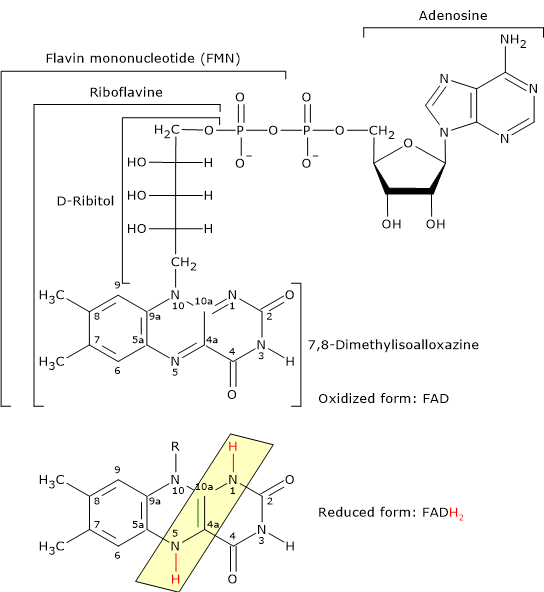
FAD is the coenzyme of dihydrolipoyl dehydrogenase, to which it is tightly bound. Like NAD, it participates in electron transfer, specifically two electrons and two protons (H+ + 2e−).[5]
Coenzyme A
Coenzyme A consists of a β-mercaptoethylamine group connected to pantothenic acid, or vitamin B5, through an amide linkage, which is in turn bonded to a 3′-phosphoadenosine moiety through a pyrophosphate bridge.
CoA participates in the reaction catalyzed by dihydrolipoyl transacetylase and acts as a carrier of acyl groups.[7]
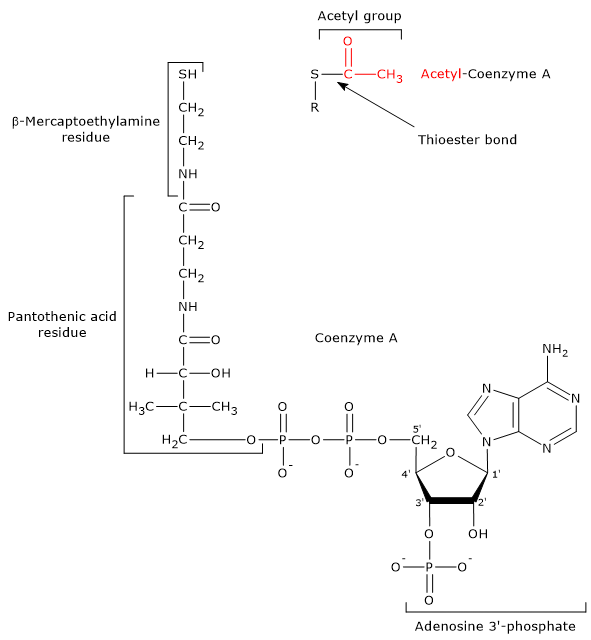
The β-mercaptoethylamine moiety terminates with a sulfhydryl group (–SH), a reactive thiol crucial to the role played by the coenzyme because acyl groups bonded to it through a thioester linkage have a high standard free energy of hydrolysis. This gives acyl groups a high transfer potential, equal to −7.5 kcal/mol (−31.5 kJ/mol), slightly more exergonic [+0.2 kcal/mol (+1 kJ/mol )] than the hydrolysis of ATP to ADP and Pi.
Therefore, thioesters have a high acyl-group transfer potential and can donate the acyl group to a variety of molecules, that is, the acyl group can be considered an activated group ready for transfer. It can also be stated that formation of the thioester bond conserves part of the free energy released during the oxidation of metabolic fuels.
Note that coenzyme A is also abbreviated as CoA-SH to emphasize the role of the thiol group.[4]
Note: in a thioester bond, a sulfur atom occupies the position where an oxygen atom is found in an ester bond.[8]
Nicotinamide adenine dinucleotide
Nicotinamide adenine dinucleotide (NAD+) can be synthesized from tryptophan, an essential amino acid, or from niacin, or vitamin B3 or vitamin PP (from “Pellagra-Preventing”), the source of the nicotinamide moiety.
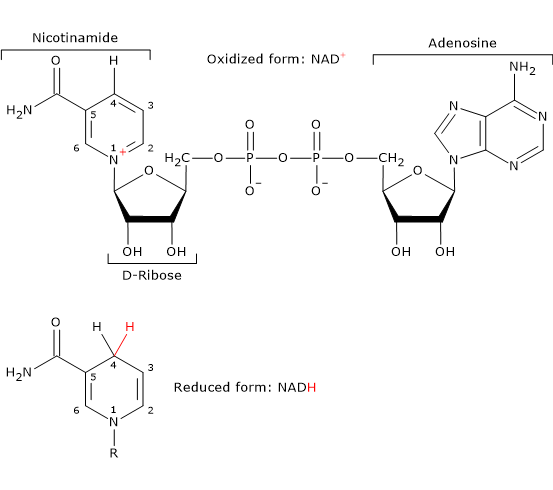
NAD+ participates in the reaction catalyzed by dihydrolipoyl dehydrogenase and, like FAD, carries out electron (hydride) transfer.[4]
Lipoic acid
Unlike the other coenzymes of the pyruvate dehydrogenase complex, lipoic acid does not derive, directly or indirectly, from vitamins and/or essential amino acids, that is, from building blocks that cannot be synthesized de novo and must be supplied through the diet.
It is the coenzyme of dihydrolipoyl transacetylase, to which it is covalently bound through an amide linkage to the ε-amino group of a lysine residue to form lipoyl-lysine, or lipoamide, the so-called lipoyl-lysine arm. It couples electron transfer with acyl-group transfer.[9]
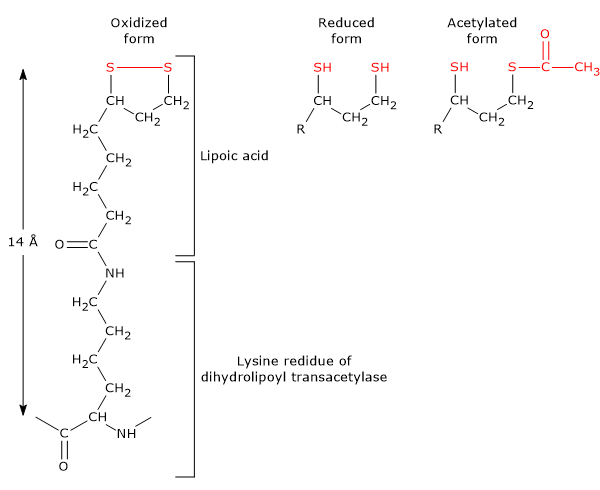
Lipoic acid has two thiol groups that can undergo reversible intramolecular oxidation to form a disulfide bridge (–S–S–), a reaction analogous to that occurring between two cysteine residues (Cys).
Because the disulfide bridge (a cyclic disulfide) can undergo redox reactions, during the reactions catalyzed by the pyruvate dehydrogenase complex it is first reduced to dihydrolipoamide, a dithiol (the reduced form of the prosthetic group), and then reoxidized to the cyclic form.[1]
Note
Many enzymes require small non-protein components, called cofactors, for their catalytic activity. Cofactors may be metal ions or small organic or organometallic molecules and are classified as coenzymes and prosthetic groups.
A prosthetic group is a cofactor that binds tightly to an enzyme by noncovalent or covalent bonds, that is, it is permanently bound to the protein. TPP, FAD, and lipoic acid are considered prosthetic groups in the PDC due to their tight binding.
A coenzyme (e.g. CoA or NAD+) is a cofactor that is not permanently bound to the enzyme.[5]
Cellular location of the pyruvate dehydrogenase complex
In eukaryotes, the pyruvate dehydrogenase complex, like the enzymes of the citric acid cycle and fatty acid oxidation, is located in the mitochondrion, where it is associated with the surface of the inner membrane facing the matrix.
In prokaryotes, it is located in the cytosol.[6]
Functions of the pyruvate dehydrogenase complex
The main functions of the pyruvate dehydrogenase complex are the production of acetyl-CoA and NADH.
The acetyl group linked to coenzyme A, an activated acetate, can be:
- oxidized to two carbon dioxide molecules via the reactions of the citric acid cycle, harvesting a portion of the potential energy in the form of ATP or GTP;
- used for the synthesis of fatty acids, cholesterol, steroids, isoprenoids, ketone bodies, and acetylcholine.
It is therefore possible to state that, depending on the metabolic conditions and/or the cell type, the pyruvate dehydrogenase complex commits carbon intermediates from amino acid and glucose catabolism to:
- the citric acid cycle, and thus to energy production, for example, in skeletal muscle under aerobic conditions, and always in cardiac muscle;
- synthesis of lipids and acetylcholine.[10]
In aerobic organisms, NADH can be oxidized to NAD+ by transferring a hydride ion to the mitochondrial electron transport chain, which in turn carries the two electrons to molecular oxygen (O2), allowing the production of 2.5 ATP molecules per pair of electrons.[1]
Note: in anaerobic organisms there are electron acceptors alternative to oxygen, such as sulfate or nitrate.[11]
Conceptually, the pyruvate dehydrogenase complex represents the bridge between glycolysis and the citric acid cycle. However, due to the irreversibility of the overall reaction catalyzed by the multienzyme complex, it acts as a one-way bridge: pyruvate can be decarboxylated and oxidized, and the remaining acetyl unit can be linked to CoA, but the reverse reaction, conversion of acetyl-CoA to pyruvate, is not possible.
The irreversibility of this reaction, together with the absence of alternative pathways, explains why acetyl-CoA, and therefore fatty acids, cannot be used as substrates for gluconeogenesis.[5]
Other sources of acetyl-CoA
Besides pyruvate, the acetyl group of acetyl-CoA can derive from the oxidation of fatty acids and the catabolism of many amino acids. However, regardless of its origin, acetyl-CoA represents an entry compound for new carbon units into the citric acid cycle. It may also be stated that the acetyl group of acetyl-CoA is the form in which most carbon enters the cycle.[10]
Mitochondrial pyruvate transport
In eukaryotes, glycolysis occurs in the cytosol, whereas all subsequent steps of aerobic metabolism, that is, the reactions catalyzed by the pyruvate dehydrogenase complex, the citric acid cycle, the electron transport chain, and oxidative phosphorylation, take place in the mitochondria.[12]
Similarly to most other metabolites and anions, the transport of pyruvate across the outer mitochondrial membrane is probably mediated by a relatively nonspecific, voltage-dependent anion channel. Conversely, its transport across the inner mitochondrial membrane occurs through a specific transporter composed of two proteins named MPC1 and MPC2 (Mitochondrial Pyruvate Carrier), which form a hetero-oligomeric complex in the membrane.[13]
Structure of the pyruvate dehydrogenase complex
Although the pyruvate dehydrogenase complex is composed of multiple copies of three different enzymes and catalyzes the same reactions through similar mechanisms in all organisms in which it is present, it nevertheless displays very different quaternary structures.[6]
Structure of the E. coli complex
The structure of E. coli multienzyme complex, which has a mass of ≈4,600 kD and a diameter of ≈300 Å, was the first to be characterized, thanks to the work of Lester Reed. In this complex, 24 units of dihydrolipoyl transacetylase form a structure with cubic symmetry; the enzymes, associated as trimers, are located at the corners of a cube. Dimers of pyruvate dehydrogenase are associated with the dihydrolipoyl transacetylase core at the center of each edge of the cube, for a total of 24 units. Finally, dimers of dihydrolipoyl dehydrogenase are located at the center of each of the six faces of the cube, for a total of 12 units. Note that the entire complex is composed of 60 units. A similar structure with cubic symmetry is also found in most other Gram-negative bacteria.[14]
Structure of the eukaryotic and Gram-positive bacterial complex
In some Gram-positive bacteria and in eukaryotes, the pyruvate dehydrogenase complex has a dodecahedral form, namely, that of a regular polyhedron with 20 vertices, 12 pentagonal faces, and 30 edges, with icosahedral symmetry (also called I symmetry). For example, the mitochondrial multienzyme complex is the largest known multienzyme complex, with a mass of ≈10,000 kDa (10 MDa) and a diameter of ≈500 Å, more than five times the size of a ribosome. Notably, it can be visualized by electron microscopy.[15][16]
The complex consists of a dodecahedral core, with a diameter of about 25 nm, formed, as in Gram-negative bacteria, by dihydrolipoyl transacetylase, but composed of 20 trimers of the enzyme (60 units) located at the vertices of the structure. The core is surrounded by 30 units of pyruvate dehydrogenase, one centered on each edge, and 12 units of dihydrolipoyl dehydrogenase, one centered on each face. The entire complex therefore contains 102 units.[2]
Additional subunits
The quaternary structure of the pyruvate dehydrgenase complex is further complicated, as mentioned previously, by the presence of three additional subunits: a pyruvate dehydrogenase kinase, a pyruvate dehydrogenase phosphatase, and the E3-binding protein (E3BP). The kinase and phosphatase are bound to the dihydrolipoyl transacetylase core.[4]
E3BP is bound to each of the 12 pentagonal faces and is therefore present in about 12 copies. It is required for the binding of dihydrolipoyl dehydrogenase to the dihydrolipoyl transacetylase core, as demonstrated by the fact that its partial proteolysis reduces the binding ability of the dehydrogenase.[17]
In the E3-binding protein, one can identify a C-terminal domain, which has no catalytic activity, and a lipoamide-containing domain, similar to that of dihydrolipoyl transacetylase, capable of accepting an acetyl group. However, removal of this domain does not reduce the catalytic activity of the multienzyme complex.[18]
Structure of pyruvate dehydrogenase
Pyruvate dehydrogenase in eukaryotes and some Gram-positive bacteria is composed of two different polypeptide chains, called α and β, which associate to form a 2-fold symmetric α2β2 heterotetramer. Conversely, in E. coli and other Gram-negative bacteria, the two subunits are fused into a single polypeptide chain, and the enzyme is therefore a homodimer.[6][19]
The enzyme contains two active sites.
In the heterotetrameric structure of Bacillus (Geobacillus) stearothermophilus pyruvate dehydrogenase, a Gram-positive bacterium, each thiamine pyrophosphate molecule binds between the N-terminal domains of an α and a β subunit, at the end of a funnel-shaped channel ≈21 Å deep that leads to the active site. Its reactive group, the thiazole ring is positioned closest to the channel entrance. At this entrance, two conserved loops are present, both essential for the catalytic activity of the enzyme and for its regulation.[20]
Active site architecture and structure
X-ray analysis of the B. stearothermophilus enzyme bound to both TPP and the peripheral subunit-binding domain (PSBD) of dihydrolipoyl transacetylase, which interacts with the C-terminal domain of the β subunits, has revealed that, in addition to having a tightly packed heterotetrameric structure, the two active sites differ structurally, particularly in the arrangement of the two conserved loops.[16] Specifically, in one enzyme subunit, in the presence of activated thiamine pyrophosphate, the inner loop is ordered in such a way that it blocks the entrance to the active site, whereas in the other subunit, the corresponding loop is disordered and does not obstruct the entrance.[21]
This observation explains, from a structural standpoint, the differences in substrate-binding rates exhibited by the two active sites. A similar arrangement and asymmetry have been observed in all thiamine pyrophosphate-dependent enzymes whose structures have been determined.
In addition to TPP and a magnesium ion (Mg2+) located in each of the two active sites, a third Mg2+ is positioned at the center of the tetramer, within a ≈20 Å deep solvent-filled tunnel that connects the two active sites. The tunnel is largely lined by ten conserved amino acid residues from all four subunits, specifically, six glutamate (Glu) and four aspartate (Asp) residues, along with other acidic residues around the TPP aminopyrimidine ring. Notably, there is a complete absence of basic residues to neutralize them. Similar tunnels have been found in all thiamine pyrophosphate-dependent enzymes with known crystal structures, whether dimeric or tetrameric, such as transketolase, an enzyme of the pentose phosphate pathway.[2]
Note: B. stearothermophilus belongs to the phylum Firmicutes and has recently been renamed Geobacillus stearothermophilus.
Role of the acidic tunnel
Mutagenesis experiments on B. stearothermophilus pyruvate dehydrogenase have shown that the tunnel plays a role in the catalytic mechanism.[22][23]
Changing some of the aforementioned acidic residues to neutral amino acids does not alter, compared with the wild-type enzyme:
- the efficiency of incorporation of the modified enzyme into the multienzyme complex;
- the structure of active sites;
- the quaternary structure of the enzyme.[24]
Nevertheless, the rate of decarboxylation is reduced by more than 70% compared with the wild-type enzyme. Likewise, once the multienzyme complex is assembled with the mutant pyruvate dehydrogenase, the overall PDC activity is reduced by more than 85% relative to the wild-type complex.[23]
How does this occur?
Because the distance between the substituted amino acids and the active sites is ≥7 Å, that is, the residues are remote from the active sites, they cannot directly influence catalytic activity. Therefore, the catalytic mechanism described below was proposed.[3]
Proton-wire mechanism and communication between the active sites
In the apoenzyme, thiamine pyrophosphate binds rapidly and tightly to the first active site, becomes activated, and the active site closes, thus protecting the zwitterionic thiazolium from the external environment.
Conversely, in the second active site TPP binds but is not activated, and the active site remains in an open conformation.[4]
In the first active site, pyruvate reacts with the thiazolium C-2. Thiamine pyrophosphate in the second active site, acting as a general acid, donates a proton to the first site. The result is decarboxylation in the first active site and activation of the coenzyme in the second site, which then closes.[3]
It should be noted that, whereas activation of the first thiamine pyrophosphate results from its binding to the active site, activation of the second coenzyme, and thus of the second active site, is coupled to the decarboxylation of pyruvate in the first active site. From another point of view: while one active site requires a general acid, the other requires a general base.[2]
Protons are required for catalytic activity, and their transport between the two active sites occurs through the acidic tunnel. They are reversibly shuttled along a chain of donor–acceptor groups provided by glutamate and aspartate residues and by the entrained water, which together act as a proton wire.
Thus, unlike many other enzymes in which communication between active sites occurs through conformational changes and subunit rearrangements, in pyruvate dehydrogenase and other TPP-dependent enzymes, the proton wire is the molecular basis for such communication.[22]
At this point, the holoenzyme has been formed, and the active sites are in dynamic equilibrium, each alternating between the dormant and activated states. This appears to be the state in which the enzyme exists in vivo at the start of each catalytic cycle.[19]
Implications of proton-wire–mediated asymmetry for catalysis
A consequence of this mechanism is that, as the catalytic cycles proceed, the two active sites remain out of phase with each other; when one active site requires a general acid, the other requires a general base, and vice versa.[22]
Finally, it should be noted that this mechanism allows the loops that close the active sites to switch in a coordinated fashion, thereby:
- coordinating substrate uptake and product release;
- explaining the asymmetry between the two active sites.[23]
Note: an apoenzyme is an enzyme lacking its cofactors. Conversely, a holoenzyme is the apoenzyme together with its cofactors. The apoenzyme is catalytically inactive, whereas the holoenzyme is catalytically active.[5]
Structure of dihydrolipoyl transacetylase
Three functionally distinct domains can be identified in the structure of dihydrolipoyl transacetylase: an N-terminal lipoyl domain, a peripheral subunit-binding domain, and a C-terminal catalytic (or acyltransferase) domain. These domains are connected by 20- to 40-residue linkers that are rich in alanine and proline, hydrophobic residues interspersed with charged ones. These linkers are highly flexible and largely extended, which allows the three domains to remain separated from each other.[25][26]
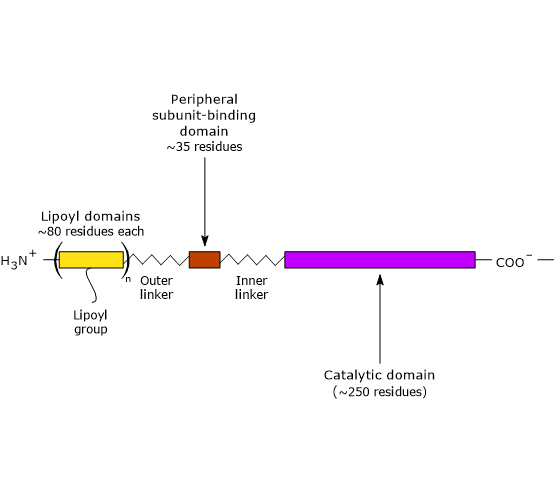
Note: flexible linkers are also present in E3BP.
The lipoyl domain
The N-terminal lipoyl domain is composed of ≈80 amino acid residues and is so called because it binds lipoic acid. The number of these domains varies by species, ranging from one to three. For example, there is one domain in Enterococcus faecalis, two in mammals, and three in Azotobacter vinelandii and E. coli.[27][28]
The linkage between the ɛ-amino group of a lysine residue and lipoic acid results in a flexible arm, the lipoyl-lysine, which has a maximum extended length of ≈14 Å. When considering the length of the lipoyl-lysine arm and the highly flexible 20- to 40-residue linker (which can span greater than 140 Å distance between active sites), the resulting flexible tether is capable of swinging the lipoyl group between the active sites of pyruvate dehydrogenase and dihydrolipoyl dehydrogenase, as well as interacting with neighboring dihydrolipoyl transacetylases within the core.[2][29]
Notably, the number of these tethers is 3 × 24 = 72 in E. coli, whereas in mammals it is 2 × 60 = 120, based on the number of N-terminal domains and the number of dihydrolipoyl transacetylase units.
A single pyruvate dehydrogenase can therefore acetylate numerous dihydrolipoyl transacetylases, and one dihydrolipoyl dehydrogenase can re-oxidize many dihydrolipoamide groups.
Moreover, the following also occur:
- interchange of acetyl groups between the lipoyl groups of the dihydrolipoyl transacetylase core;
- exchange of both acetyl groups and disulfides between the tethered arms.[25]
The peripheral subunit-binding domain
PSBD is composed of ≈35 amino acid residues arranged into a globular structure that binds both pyruvate dehydrogenase and dihydrolipoyl dehydrogenase; that is, it helps hold the multienzyme complex together.[30]
The C-terminal catalytic domain
The C-terminal catalytic domain, which contains the active site, consists of ≈250 amino acid residues arranged into a hollow, cage-like structure containing channels large enough to allow substrates and products to diffuse in and out. For example, CoA and lipoamide, the two substrates of dihydrolipoyl transacetylase, bind in their extended conformations at opposite ends of a channel located at the interface between each pair of subunits in each trimer.[26][31]
Structure of dihydrolipoyl dehydrogenase
The structure of dihydrolipoyl dehydrogenase has been deduced from studies of the enzyme in several microorganisms. It has a homodimeric structure, with each ≈470-residue polypeptide chain folded into four domains, from the N-terminal to the C-terminal end: an FAD-binding domain, a NAD+-binding domain, a central domain, and an interface domain. All domains participate in the formation of the active site.[32]
FAD is almost completely buried inside the protein because, unlike a thiol or NADH, it is easily oxidized and must therefore be protected from the surrounding solution. In fact, in the absence of the nicotinamide coenzyme, the phenolic side chain of a tyrosine residue (e.g. Tyr181 in the Gram-negative bacterium Pseudomonas putida), covers the NAD+-binding pocket so as to protect FADH2 from exposure to the solvent.[2]
Conversely, when NAD+ occupies the active site, the phenolic side chain of the aforementioned tyrosine residue becomes interposed between the nicotinamide ring and the flavin ring.[33]
In the active site of the enzyme’s oxidized form, a redox-active disulfide bridge is also present. It forms between two cysteine residues located in a highly conserved segment of the polypeptide chain, e.g., Cys43 and Cys48 in P. putida, and is positioned on the side of the flavin ring opposite the nicotinamide ring. The disulfide bridge connects consecutive turns of a distorted α-helix; notably, without such distortion, the Cα atoms of the two cysteine residues would be too far apart to allow disulfide bond formation.[34]
Dihydrolipoyl dehydrogenase therefore has two electron acceptors: FAD and the redox-active disulfide bridge.
Note: the heterocyclic rings of NAD and FAD are parallel and in contact through van der Waals interactions; S48 is also in van der Waals contact with the flavin ring on the side opposite the NAD ring.[2]
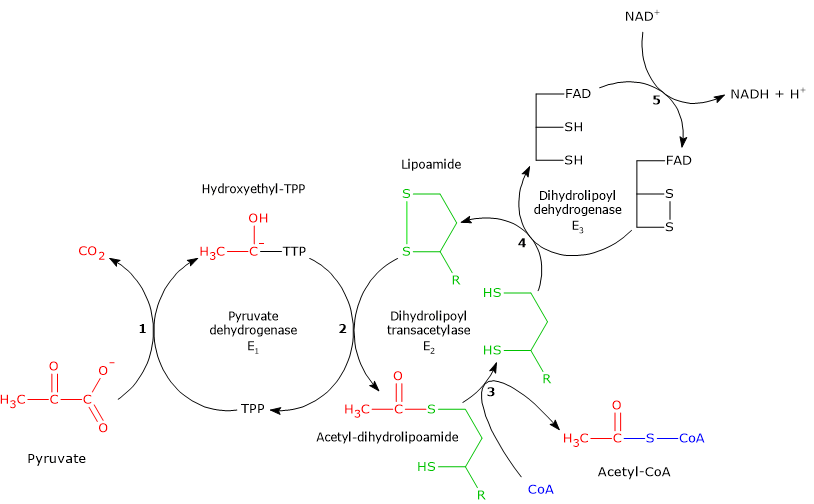
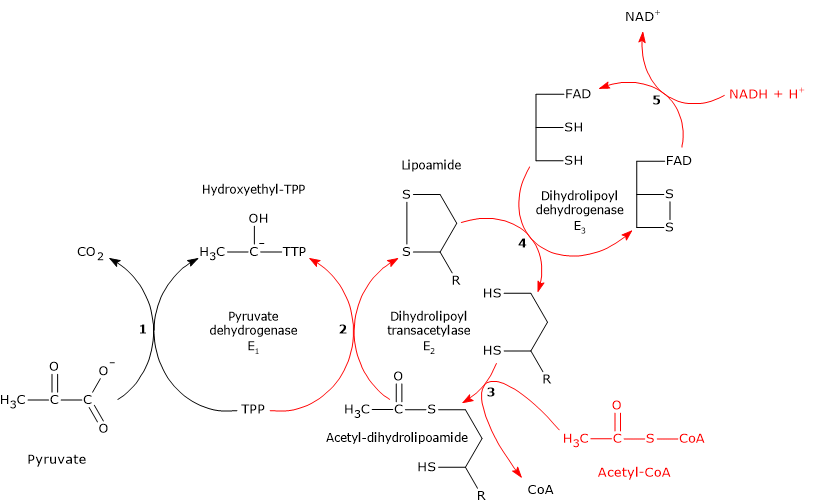
Reaction of pyruvate dehydrogenase
In the reaction sequence catalyzed by the components of the pyruvate dehydrogenase complex, pyruvate dehydrogenase catalyzes the first two steps:
- the decarboxylation of pyruvate to form CO2 and the hydroxyethyl-TPP intermediate;
- the reductive acetylation of the lipoyl group of dihydrolipoyl transacetylase.
The first reaction is essentially identical to the reaction catalyzed by pyruvate decarboxylase (EC 4.1.1.1), which carries out a non-oxidative decarboxylation during glucose fermentation to ethanol. What differs is the fate of the hydroxyethyl group bound to thiamine pyrophosphate: in the reaction catalyzed by pyruvate dehydrogenase, this group is transferred to the next enzyme in the sequence, dihydrolipoyl transacetylase, whereas in the reaction catalyzed by pyruvate decarboxylase it is converted into acetaldehyde.[4]
Catalytic mechanism: the first stage
In thiamine pyrophosphate-dependent enzymes, the thiazolium ring is the active center, but only in its dipolar carbanion, or ylid, form, that is, a zwitterion (German for “hybrid ion”) with a positive charge on N-3 and a negative charge on C-2. Conversely, the positively charged thiazolium ring (positively charged nitrogen and no charge on C-2) can be considered the “dormant” or inactive form.[35]
The reaction begins with the nucleophilic attack by the C-2 carbanion on the carbonyl carbon of pyruvate, which has the oxidation state of an aldehyde, leading to the formation of a covalent bond between the coenzyme and pyruvate.
Then, cleavage of the C-1–C-2 bond of pyruvate occurs. This results in the release of the carboxyl group, namely C-1 as CO2, while the remaining carbon atoms, C-2 and C-3, remain bound to thiamine pyrophosphate as a hydroxyethyl group. Cleavage of the C-1–C-2 bond, and thus pyruvate decarboxylation, is favored because the negative charge on carbon C-2, which would otherwise be unstable, is stabilized by the positively charged N-3 of the thiazolium ring, an iminium nitrogen (C=N+). This electrophilic, electron-deficient structure acts as an electron sink, in which the carbanion electrons can be delocalized by resonance.[36]
At this point, the resonance-stabilized intermediate can be protonated to form hydroxyethyl–TPP.[5]
Note: this first reaction catalyzed by pyruvate dehydrogenase is the step at which the pyruvate dehydrogenase complex exerts its substrate specificity; furthermore, it is the slowest of the five reactions and thus rate-limiting for the overall process.[1]
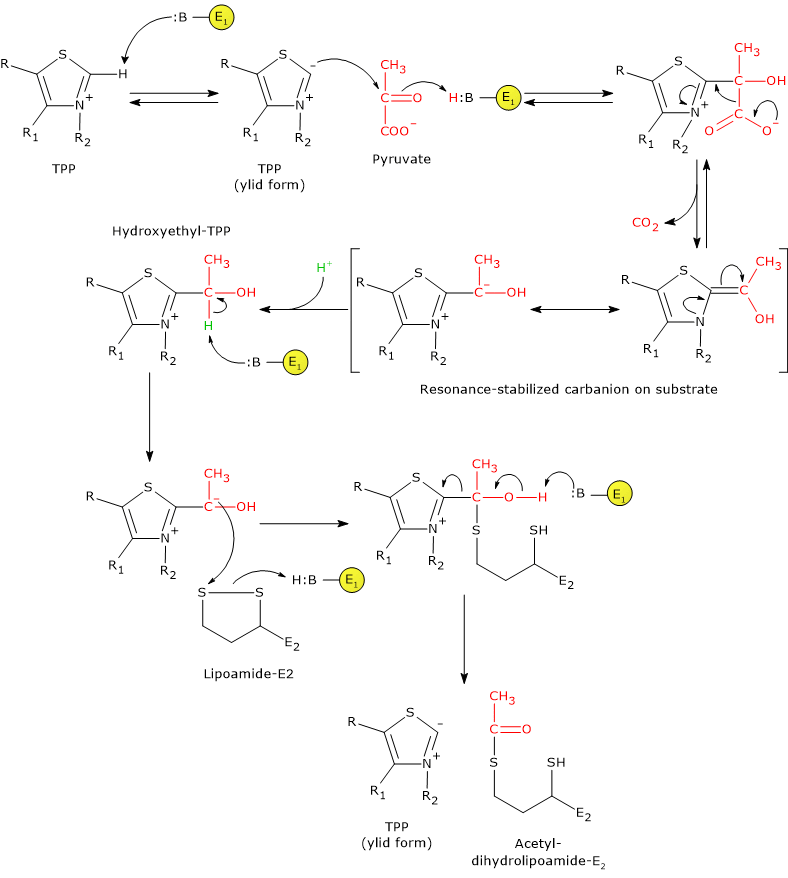
Catalytic mechanism: the second stage
Pyruvate dehydrogenase then catalyzes the oxidation of the hydroxyethyl group to an acetyl group and its transfer to the lipoyl-lysyl arm of dihydrolipoyl transacetylase. The reaction begins with the formation of a carbanion on the hydroxyl-bearing carbon of hydroxyethyl–TPP, via removal of its proton by an enzyme base.[5]
The carbanions carry out a nucleophilic attack on the lipoamide disulfide, forming a high-energy acetyl-thioester bond with one of the two –SH groups. During this reaction, oxidation of the hydroxyethyl group to an acetyl group occurs simultaneously with reduction of the lipoamide disulfide bond: the two electrons removed from the hydroxyethyl group are used to reduce the disulfide. This reaction is therefore a reductive acetylation accompanied by regeneration of the active form of pyruvate dehydrogenase, namely the enzyme with thiazolium C-2 in its deprotonated, ylid (dipolar carbanion) form.[36]
Note that the energy derived from the oxidation of the hydroxyethyl group to an acetyl group drives the formation of the thioester bond between the acetyl group and lipoamide.[4]
Note: as previously mentioned, the lipoyl-lysyl arm, arranged in an extended conformation and able to access the active site of pyruvate dehydrogenase where TPP is bound, enables the transfer of the acetyl group, generated by oxidation of hydroxyethyl–TPP, to the lipoamide moiety. The swinging arm then delivers the acetyl group to the active site of dihydrolipoyl transacetylase for transfer to coenzyme A. It can subsequently move to the active site of dihydrolipoyl dehydrogenase.[2]
A deeper look on thiamine pyrophosphate
Thiamine pyrophosphate consists of three chemical moieties on which its chemistry and enzymology depend: a thiazolium ring, a 4-aminopyrimidine ring, and the diphosphate side chain.[37]
The diphosphate side chain anchors the cofactor to the enzyme through electrostatic interactions between its negatively charged phosphoryl groups and the positively charged Ca2+ and Mg2+ ions, which are themselves bound to highly conserved sequences, Gly-Asp-Gly (GDG) and GlyAspGly-X26-AsnAsn (GDG-X26-NN), respectively.[38]
The thiazolium ring plays the central role in catalysis because of its ability to form the C-2 carbanion, which acts as a nucleophilic center on the C-2 atom.[39]
Note: as mentioned previously, once bound to the enzyme, thiamine pyrophosphate positions itself in the active site so that the thiazolium ring lies near the entrance of the channel leading to the active site.[5]
The aminopyrimidine ring has a dual function:
- it anchors the coenzyme, holding it in place;
- it has a specific catalytic role, participating in acid–base catalysis, as demonstrated by studies with TPP analogs in which the three nitrogen atoms of the ring were substituted in turn. These studies showed that the N-1′ atom and the N-4′ amino group are essential, whereas the N-3′ atom is required to a lesser extent.[4]
How the dipolar carbanion of thiamine pyrophosphate is formed
Three tautomeric forms of the aminopyrimidine ring can be identified in the enzyme-bound coenzyme (before substrate entry):
- the canonical 4’-aminopyrimidine tautomer;
- the N-1 protonated form (the 4-aminopyrimidinium ion);
- the 1’,4’-iminopyrimidine tautomer.[40]
Evidence indicates that the 1′,4′-imino tautomer is the form that undergoes deprotonation before the substrate enters the active site. The C-2 proton of the thiazolium ring is “much more acidic than most =C–H groups in other molecules.” Its unusually high acidity, i.e., the ease with which the C-2 proton dissociates, is due to the presence of the quaternary nitrogen in the thiazolium ring, a positively charged atom capable of stabilizing the resulting carbanion by electrostatic attraction.[41]
In the deprotonation reaction, the amino group of the aminopyrimidine ring appears to play an essential role: it acts as a base and is ideally positioned to accept the proton. However, in the 4′-aminopyrimidine tautomer, one of its protons sterically clashes with the C-2 proton, and its pKa is too low to perform the deprotonation efficiently.[37]
A mechanism was therefore proposed in which the side chain of a conserved glutamate residue, for example, βGlu59 in Bacillus stearothermophilus or Glu51 in Saccharomyces uvarum (brewer’s yeast) pyruvate decarboxylase, donates a proton to the aminopyrimidine ring. This converts it to the 1′,4′-iminopyrimidine tautomer, which then accepts the C-2 proton and reverts to the canonical 4′-aminopyrimidine form, enabling formation of the carbanion.[2]
Note: formation of the C-2 carbanion occurs through an intramolecular proton transfer.
Deprotonation of thiamine pyrophosphate
Loss of the C-2 proton of the thiazolium ring converts it from a positively charged ring into a dipolar ion (zwitterion). This change in charge state induces a conformational change in one of the two conserved loops located at the entrance of the active site channel, specifically the inner loop, which in turn causes the channel to close to the surrounding aqueous environment. In this closed conformation, the thiazolium carbanion is protected from electrophiles.[42]
To summarize: deprotonation of thiamine pyrophosphate leads to closure of the active site and protection of the newly formed dipolar carbanion. Thus, TPP-dependent enzymes are active only in the closed conformation.
Conversely, in the other active site (since these enzymes typically have two active sites per dimer), thiamine pyrophosphate is not in the ylid form, the channel remains open, and that site is inactive.[5]
Reaction of dihydrolipoyl transacetylase
In the reaction sequence catalyzed by the components of the pyruvate dehydrogenase complex, dihydrolipoyl transacetylase catalyzes the third step: the transfer of the acetyl group from acetyl-dihydrolipoamide to CoA to form acetyl-CoA and dihydrolipoamide, the fully reduced dithiol form of lipoamide.
It should be noted that the acetyl group, initially linked by a tioester bond to one of the –SH groups of lipoamide, is subsequently transferred to the –SH group of coenzyme A, also via an tioester bond, hence the term transtioesterification.[4]
Catalytic mechanism
During the reaction, the sulfhydryl group of coenzyme A performs a nucleophilic attack on the carbonyl carbon of the acetyl group of acetyl-dihydrolipoamide (bound to dihydrolipoyl transacetylase). This forms a transient tetrahedral intermediate, which then decomposes to produce dihydrolipoamide–dihydrolipoyl transacetylase and acetyl-CoA.
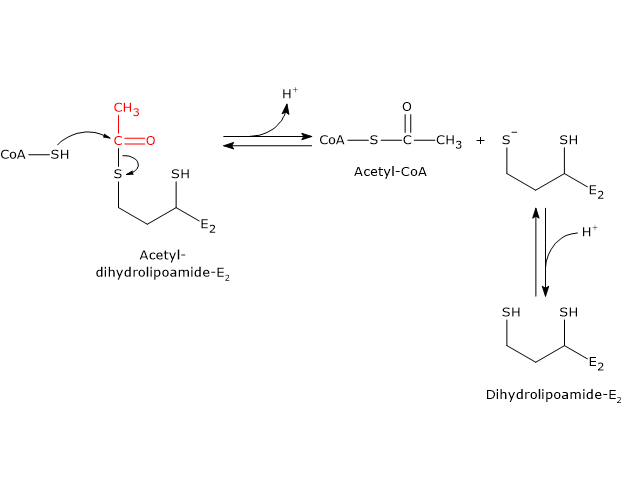
As mentioned earlier, the mobility of the lipoyl-lysyl arm plays a central role in this mechanism.[43][44]
Reaction of dihydrolipoyl dehydrogenase
In the reaction sequence catalyzed by the pyruvate dehydrogenase complex, dihydrolipoyl dehydrogenase catalyzes the fourth and fifth steps. The enzyme mediates the electron transfers required to regenerate the disulfide bridge of the lipoyl group of dihydrolipoyl transacetylase, that is, to restore the oxidized form of the prosthetic group, thus completing the catalytic cycle of the transacetylase.[5]
The reaction follows a ping-pong catalytic mechanism: it proceeds through two successive half-reactions in which each of the two substrates, NAD⁺ and dihydrolipoamide, reacts in the absence of the other. During the first half-reaction, release of the first product and formation of an enzyme intermediate occur before the second substrate binds, and the enzyme undergoes a structural change. During the second half-reaction, release of the second product occurs, and the enzyme returns to its initial state, again through a conformational change.[2]
According to the ping-pong kinetic mechanism:
- in the first half-reaction, dihydrolipoamide is oxidized to lipoamide;
- in the second half-reaction, NAD+ is reduced to NADH.[4][45]
Catalytic mechanism: first half-reaction
Below, the reaction mechanism of P. putida dihydrolipoyl dehydrogenase is described.
In the first half-reaction, oxidized dihydrolipoyl dehydrogenase (E), i.e., the enzyme containing the Cys43–Cys48 disulfide, binds dihydrolipoamide (LH2) to form the enzyme–dihydrolipoamide complex (E–LH₂). Subsequently, a sulfur atom of dihydrolipoamide performs a nucleophilic attack a nucleophilic attack on the sulfur of Cys43, forming the lipoamide–Cys43 disulfide bridge (E–S–S–L), while the sulfur of Cys48 is released as a thiolate ion (S48−).[34]
The proton on the second thiol group of lipoamide is then abstracted by histidine (His) 451, which acts as a general acid–base catalyst, leading to the formation of a second thiolate ion, this time on the lipoamide (E–S–S–L–S⁻). This thiolate, through a nucleophilic attack, displaces the sulfur of Cys43, S43, aided by general acid catalysis from His451, which donates a proton to S43. The catalytic action of Hys451 is essential, as demonstrated by mutagenesis studies in which its substitution with a glutamine residue causes the enzyme to retain ≈0.4% of the wild-type catalytic activity.[18]
Then, the thiolate anion S48− interacts, through non-covalent contacts, with the flavin ring near the 4a position; that is, an electron pair of S48−, acting as electron donor, is partially transferred to the oxidized flavin ring, which acts as the electron acceptor. The resulting structure is called a charge-transfer complex.
Meanwhile, the phenolic side chain of Tyr181 continues to hinder access to the flavin ring, thereby protecting it from oxidation by O2.[2]
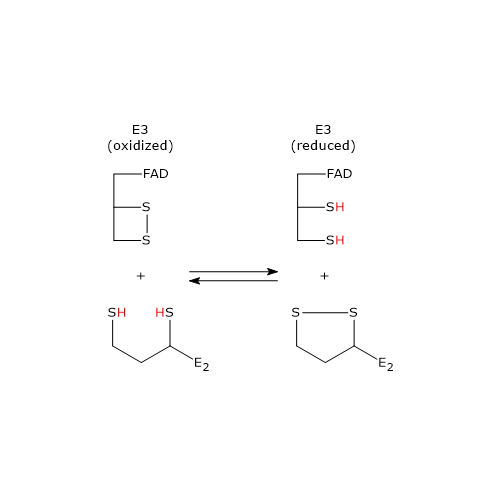
To summarize, what occurs is an interchange reaction of disulfide bridges leading to the formation of the oxidized form of lipoamide, the first product, which is released, and the reduced form of dihydrolipoyl dehydrogenase.
Catalytic mechanism: second-half reaction
The second half-reaction involves the reduction of NAD+ to NADH + H+ by electron transfer from the reactive disulfide of the enzyme via FAD.
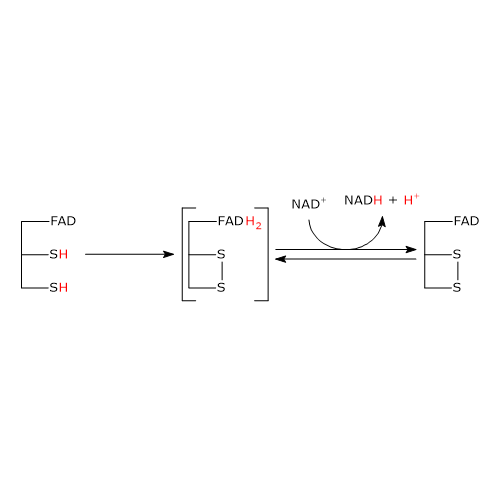
It begins with the entry of NAD+ into the active site and its binding to form the E−H2−NAD+ complex. It should be noted that the entry of the coenzyme causes the phenolic side chain of Tyr181 to be pushed aside by the nicotinamide ring.
Following the collapse of the charge-transfer complex, a covalent bond is formed between flavin atom C-4a and S48, accompanied by the extraction of a proton from S43 by flavin atom N-5, yielding the corresponding thiolate anion (S43−).
S43− then carries out a nucleophilic attack on S48, leading to the formation of the redox-active disulfide bridge between Cys43 and Cys48, followed by the cleavage of the covalent bond between S48 and flavin atom C-4a, producing the reduced FADH− anion with a negative charge on atom N-1.
It should be noted that dihydrolipoyl dehydrogenase is now in the oxidized form (E).[34]
FADH− has a transient existence because it instantly transfers a hydride ion to nicotinamide atom C-4, which is juxtaposed to flavin atom N-5. This results in the formation of FAD and of the second product of the reaction, NADH, which is released.[1]
Functional implications and comparison with glutathione reductase
To summarize, what occurs is that the electrons removed from the hydroxyethyl group, which derives from pyruvate, pass via FAD to NAD+. The catalytic cycle of dihydrolipoyl dehydrogenase is therefore completed, with the enzyme and its coenzymes in their oxidized forms. At this point, the catalytic cycle of the entire pyruvate dehydrogenase complex is also completed, and the complex is ready for a new reaction cycle.[1]
Note: unlike the thiazolium ring of thiamine pyrophosphate, FAD does not act as an electron trap or electron sink, but rather as an electron conduit between the reduced redox-active disulfide and NAD+.
The catalytic mechanism of dihydrolipoyl dehydrogenase has been determined by analogy with that of glutathione reductase (EC 1.8.1.7), which is 33% identical and whose structure is more extensively characterized.[46] It should, however, be noted that although the two enzymes catalyze similar reactions, these generally occur in opposite directions:
- dihydrolipoyl dehydrogenase uses NAD+ to oxidize two –SH groups to a disulfide (–S–S–);
- glutathione reductase uses NADPH to reduce a –S–S– to two thiol groups.
Nevertheless, their active sites are closely superimposable.[2]
Regulation of the pyruvate dehydrogenase complex
In mammals, the regulation of the activity of the pyruvate dehydrogenase complex is essential in both the fed and fasted states. In fact, this multienzyme complex plays a central role in metabolism because, by catalyzing the irreversible oxidative decarboxylation of pyruvate, it represents the entry point of carbon flux from all carbohydrate sources as well as from ≈50% of the carbon skeletons of glucogenic amino acids, which, taken together, account for ≈60% of daily caloric intake, into:
- the citric acid cycle, and therefore to full oxidation to CO2;
- the synthesis of lipids (fed state) and acetylcholine.[47]
The importance of regulating the conversion of pyruvate into acetyl-CoA is also underscored by the fact that mammals, although able to produce glucose from pyruvate, cannot synthesize it from acetyl-CoA because of the irreversibility of the pyruvate dehydrogenase reaction and the absence of alternative pathways. Thus, inhibition of the activity of the complex allows glucose, and amino acids that can be converted into pyruvate, such as alanine, to be spared when other fuels, for example acetyl-CoA from fatty acid oxidation, are available.[48]
This explains why the activity of the complex is tightly regulated by:
- feedback inhibition;
- nucleotides;
- covalent modifications, namely phosphorylation and dephosphorylation of specific target proteins.[5]
Regulation by feedback inhibition and the energy status of the cell
The activity of the dephosphorylated form of the pyruvate dehydrogenase complex is regulated by feedback inhibition.
Acetyl-CoA and NADH allosterically inhibit the enzymes that catalyze their formation, dihydrolipoyl transacetylase and dihydrolipoyl dehydrogenase, respectively.[49]
In addition, CoA and acetyl-CoA, as well as NAD+ and NADH, compete for binding sites on E2 and E3, respectively, which catalyze reversible reactions. This means that, when the ratios [acetyl-CoA]/[CoA] and [NADH]/[NAD+] are high, the transacetylation and dehydrogenation reactions proceed in reverse. Consequently, dihydrolipoyl transacetylase cannot accept the hydroxyethyl group from TPP because it is maintained in the acetylated form. This causes thiamine pyrophosphate to remain bound to pyruvate dehydrogenase in its hydroxyethyl form, which, in turn, decreases the rate of pyruvate decarboxylation. Hence, high [acetyl-CoA]/[CoA] and [NADH]/[NAD+] ratios indirectly influence pyruvate dehydrogenase activity.[1]
Links with fatty acid oxidation
Acetyl-CoA and NADH are also produced by fatty acid oxidation, which, like the reactions of the pyruvate dehydrogenase complex, takes place within the mitochondrion. This means that the cell, by regulating the activity of the multienzyme complex, preserves carbohydrate stores when fatty acids are available for energy. For example, during the fasted state, the liver, skeletal muscle, and many other organs and tissues rely primarily on fatty acid oxidation for energy. Conversely, the activity of the multienzyme complex is increased in the fed state, when many different types of cells and tissues mainly use glucose as a fuel.
More generally, when the production of NADH and/or acetyl-CoA exceeds the capacity of the cell to use them for ATP production, the activity of the pyruvate dehydrogenase complex is inhibited. The same is true when there is no need for additional ATP to be produced. In fact, the activity of the multienzyme complex is also sensitive to the energy charge of the cell. Through allosteric mechanisms, high ATP levels inhibit the activity of the pyruvate dehydrogenase component of the complex, whereas high ADP levels, which signal that the energy charge of the cell may be falling, activate it, thus committing the carbon skeletons of carbohydrates and some amino acids to energy production.
Note: in skeletal muscle, the activity of the pyruvate dehydrogenase complex increases with increased aerobic activity, resulting in a greater dependence on glucose as a fuel source.[4]
Regulation by phosphorylation/dephosphorylation
Unlike prokaryotes, in mammals the activity of the pyruvate dehydrogenase complex is also regulated by covalent modifications, namely, the phosphorylation and dephosphorylation of three specific serine residues on the α-subunit of pyruvate dehydrogenase, the enzyme that catalyzes the first and irreversible step of the overall reaction sequence.
Note: since mammalian pyruvate dehydrogenase is a heterotetramer, there are six potential phosphorylation sites.[1]
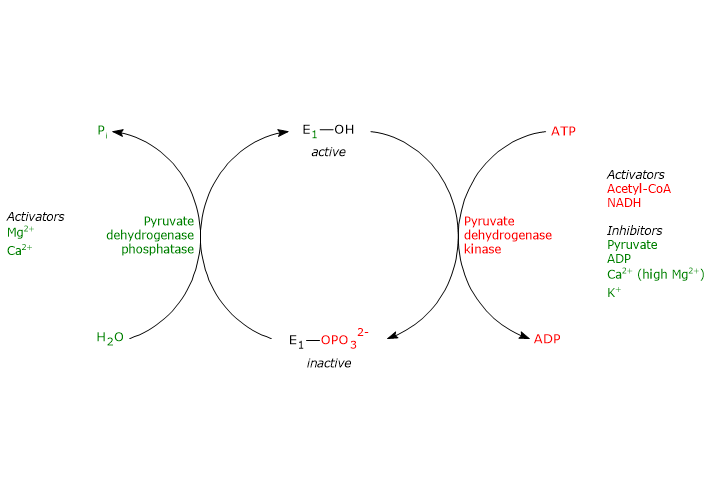
Phosphorylation, which inactivates pyruvate dehydrogenase and thereby blocks the overall reaction sequence, is catalyzed by pyruvate dehydrogenase kinase. Two of the aforementioned serine residues are located on the more C-terminal loop at the entrance of the substrate channel leading to the respective active site, and phosphorylation of only one of them is sufficient to inactivate pyruvate dehydrogenase, thus demonstrating the out-of-phase coupling between its active sites.[50][51]
Conversely, in the dephosphorylated state the complex is active. Dephosphorylation is catalyzed by a specific protein phosphatase, pyruvate dehydrogenase phosphatase.[52]
The activities of pyruvate dehydrogenase kinase and pyruvate dehydrogenase phosphatase are, in turn, subject to allosteric regulation by several modulators.[53]
Regulation of pyruvate dehydrogenase kinase
The activity of pyruvate dehydrogenase kinase depends on the ratios of [NADH]/[NAD+], [acetyl-CoA]/[CoA], and [ATP]/[ADP], as well as on the pyruvate concentration in the mitochondrial matrix.[54]
- High ratios of [NADH]/[NAD+] and [acetyl-CoA]/[CoA], such as those occurring during the oxidation of fatty acids and ketone bodies, activate the kinase. As a result, pyruvate dehydrogenase is phosphorylated, and the pyruvate dehydrogenase complex is inhibited. This allows tissues, such as cardiac muscle, to preserve glucose when fatty acids and/or ketone bodies are being used for energy, because acetyl-CoA synthesis from pyruvate, and therefore from carbohydrates (and some amino acids), is turned off. Conversely, when the concentrations of NAD+ and coenzyme A are high, the activity of the kinase is inhibited and the multienzyme complex is active. Therefore, acetyl-CoA and NADH, two of the three end products of the reactions catalyzed by the pyruvate dehydrogenase complex, control their own synthesis allosterically, both directly and indirectly, by regulating the activity of pyruvate dehydrogenase kinase.
- High ratio of [ATP]/[ADP] activates the kinase and thereby inhibits the pyruvate dehydrogenase complex. Note: unlike many other kinases, such as those involved in regulating glycogen metabolism, pyruvate dehydrogenase kinase is not regulated by cAMP levels, but rather by molecules that signal changes in the energy status of the cell and the availability of biosynthetic intermediates, ATP and NADH, and acetyl-CoA, respectively.
- Pyruvate allosterically inhibits pyruvate dehydrogenase kinase. When its levels are high, it binds to the kinase and inactivates it; pyruvate dehydrogenase is not phosphorylated, and the pyruvate dehydrogenase complex remains active.
- Pyruvate dehydrogenase kinase is also activated by interaction with dihydrolipoyl transacetylase in its acetylated form, i.e., when acetyl-dihydrolipoamide is present.
Other activators of the kinase is potassium and magnesium ions.[2][53]
Regulation of pyruvate dehydrogenase phosphatase
The activity of pyruvate dehydrogenase phosphatase depends on the ratios of [NADH]/[NAD+] and [acetyl-CoA]/[CoA], as well as on the concentration of Ca2+ in the mitochondrial matrix.
- Low ratios of [NADH]/[NAD+] and [acetyl-CoA]/[CoA] activate the phosphatase; pyruvate dehydrogenase is dephosphorylated, and the pyruvate dehydrogenase complex is activated. Conversely, when these ratios are high, phosphatase activity decreases, kinase activity increases, and the multienzyme complex is inhibited.
- Calcium ion activates pyruvate dehydrogenase phosphatase.
Ca2+ s an important second messenger that signals that the cell requires more energy. Therefore, when its levels are high, as in cardiac muscle cells after epinephrine stimulation or in skeletal muscle cells during muscular contraction, the phosphatase is active, the complex is dephosphorylated, and thus active. - Insulin, also contributes to the regulation of the pyruvate dehydrogenase complex by activating pyruvate dehydrogenase phosphatase. In response to increases in blood glucose, the hormone stimulates glycogen synthesis and the synthesis of acetyl-CoA, a precursor for lipid biosynthesis.[2][53]
Fasting and subsequent refeeding also affect the activity of the multienzyme complex.
In tissues such as skeletal muscle, cardiac muscle, and kidney, fasting significantly decreases the activity of the complex, whereas refeeding reverses the inhibition produced by fasting. In the brain, however, these variations are not observed, because the activity of the pyruvate dehydrogenase complex is essential for ATP production.[1][55]
References
- ^ a b c d e f g h i l Nelson D.L., Cox M.M. Lehninger. Principles of biochemistry. 8th Edition. W.H. Freeman and Company, 2021.
- ^ a b c d e f g h i j k l m n o Heilman D., Woski S., Voet D., Voet J.G., Pratt C.W. Fundamentals of biochemistry: life at the molecular level. 6th Edition. Wiley, 2023.
- ^ a b c Patel M.S., Nemeria N.S., Furey W., Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem 2014;289(24):16615-16623. doi:10.1074/jbc.R114.563148
- ^ a b c d e f g h i j k Berg J.M., Tymoczko J.L., Gregory J.G. Jr, Stryer L. Biochemistry. 9th Edition. W.H. Freeman and Company, 2019.
- ^ a b c d e f g h i j k Garrett R.H., Grisham C.M. Biochemistry. 6th Edition. Brooks/Cole, Cengage Learning, 2016.
- ^ a b c d Bothe S.N., Zdanowicz R. Structural diversity of pyruvate dehydrogenase complexes. FEBS Lett 2025. doi:10.1002/1873-3468.70140
- ^ Czumaj A., Szrok-Jurga S., Hebanowska A., Turyn J., Swierczynski J., Sledzinski T., Stelmanska E. The pathophysiological role of CoA. Int J Mol Sci 2020;21(23):9057. doi:10.3390/ijms21239057
- ^ Solomons T.W.G., Fryhle C.B., Snyder S.A. Solomons’ organic chemistry. 12th Edition. John Wiley & Sons Incorporated, 2017.
- ^ Solmonson A., DeBerardinis R.J. Lipoic acid metabolism and mitochondrial redox regulation. J Biol Chem 2018;293(20):7522-7530. doi:10.1074/jbc.TM117.000259
- ^ a b Stipanuk M.H., Caudill M.A. Biochemical, physiological, and molecular aspects of human nutrition. 4th Edition. St. Louis: Elsevier, 2018.
- ^ Lu Z., Imlay J.A. When anaerobes encounter oxygen: mechanisms of oxygen toxicity, tolerance and defence. Nat Rev Microbiol 2021;19(12):774-785. doi:10.1038/s41579-021-00583-y
- ^ Chaudhry R., Varacallo M.A. Biochemistry, Glycolysis. [Updated 2023 Aug 8]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK482303/
- ^ Zangari J., Petrelli F., Maillot B., Martinou J.C. The multifaceted pyruvate metabolism: role of the mitochondrial pyruvate carrier. Biomolecules 2020;10(7):1068. doi:10.3390/biom10071068
- ^ Reed L.J., Pettit F.H., Eley M.H., Hamilton L., Collins J.H., Oliver R.M. Reconstitution of the Escherichia coli pyruvate dehydrogenase complex. Proc Natl Acad Sci USA. 1975;72(8):3068-72. doi:10.1073/pnas.72.8.3068
- ^ Stoops J.K., Cheng R.H., Yazdi M.A., et al. On the unique structural organization of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. J Biol Chem 1997;272(9):5757-64. doi:10.1074/jbc.272.9.5757
- ^ a b Zhou Z.H., McCarthy D.B., O’Connor C.M., Reed L.J., Stoops J.K. The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc Natl Acad Sci USA 2001;98(26):14802-7. doi:10.1073/pnas.011597698
- ^ Forsberg B.O., Aibara S., Howard R.J., Mortezaei N., Lindahl E. Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex. Nat Commun 2020;11(1):4667. doi:10.1038/s41467-020-18401-z
- ^ a b Ciszak E.M., Makal A., Hong Y.S., Vettaikkorumakankauv A.K., Korotchkina L.G., Patel M.S. How dihydrolipoamide dehydrogenase-binding protein binds dihydrolipoamide dehydrogenase in the human pyruvate dehydrogenase complex. J Biol Chem 2006;281(1):648-55. doi:10.1074/jbc.M507850200
- ^ a b Wang C., Ma C., Xu Y., et al. Dynamics of the mammalian pyruvate dehydrogenase complex revealed by in-situ structural analysis. Nat Commun 2025;16(1):917. doi:10.1038/s41467-025-56171-8
- ^ Milne J.L., Wu X., Borgnia M.J., Lengyel J.S., Brooks B.R., Shi D., Perham R.N., Subramaniam S. Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J Biol Chem 2006;281(7):4364-70. doi:10.1074/jbc.M504363200
- ^ Kale S., Arjunan P., Furey W., Jordan F. A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase complex E1 component modulates substrate utilization and chemical communication with the E2 component. J Biol Chem 2007;282(38):28106-16. doi:10.1074/jbc.M704326200
- ^ a b c Frank R.A.W., Titman C.M., Pratap J.V., Luisi B.F., Perham R.N. A molecular switch and proton wire synchronize the active sites in thiamine enzymes. Science 2004;306(5697):872-876. doi:10.1126/science.1101030
- ^ a b c Nemeria N.S., Arjunan P., Chandrasekhar K., Mossad M., Tittmann K., Furey W., Jordan F. Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a “direct pathway” between the 4′-aminopyrimidine N1′ atoms. J Biol Chem 2010;285(15):11197-209. doi:10.1074/jbc.M109.069179
- ^ Arjunan P., Nemeria N., Brunskill A., Chandrasekhar K., Sax M., Yan Y., Jordan F., Guest J.R., Furey W. Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution. Biochemistry 2002;41(16):5213-21. doi:10.1021/bi0118557
- ^ a b Perham R.N. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem 2000;69:961-1004. doi:10.1146/annurev.biochem.69.1.961
- ^ a b Wang J., Nemeria N.S., Chandrasekhar K., et al. Structure and function of the catalytic domain of the dihydrolipoyl acetyltransferase component in Escherichia coli pyruvate dehydrogenase complex. J Biol Chem 2014;289(22):15215-30. doi:10.1074/jbc.M113.544080
- ^ Allen A.G., Perham R.N. Two lipoyl domains in the dihydrolipoamide acetyltransferase chain of the pyruvate dehydrogenase multienzyme complex of Streptococcus faecalis. FEBS Lett 1991;287(1-2):206-10. doi:10.1016/0014-5793(91)80052-5
- ^ Hanemaaijer R., de Kok A., Jolles J., Veeger C. The domain structure of the dihydrolipoyl transacetylase component of the pyruvate dehydrogenase complex from Azotobacter vinelandii. Eur J Biochem 1987;169(2):245-52. doi:10.1111/j.1432-1033.1987.tb13604.x
- ^ Hale G., Perham R.N. Amino acid sequence around lipoic acid residues in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem J 1980;187(3):905-8. doi:10.1042/bj1870905
- ^ Hipps D.S., Packman L.C., Allen M.D., Fuller C., Sakaguchi K., Appella E., Perham R.N. The peripheral subunit-binding domain of the dihydrolipoyl acetyltransferase component of the pyruvate dehydrogenase complex of Bacillus stearothermophilus: preparation and characterization of its binding to the dihydrolipoyl dehydrogenase component. Biochem J 1994;297(Pt 1):137-43. doi:10.1042/bj2970137
- ^ Schulze E., Westphal A.H., Obmolova G., Mattevi A., Hol W.G., de Kok A. The catalytic domain of the dihydrolipoyl transacetylase component of the pyruvate dehydrogenase complex from Azotobacter vinelandii and Escherichia coli. Expression, purification, properties and preliminary X-ray analysis. Eur J Biochem 1991;201(3):561-8. doi:10.1111/j.1432-1033.1991.tb16315.x
- ^ Duarte I.F., Caio J., Moedas M.F., Rodrigues L.A., Leandro A.P., Rivera I.A., Silva M.F.B. Dihydrolipoamide dehydrogenase, pyruvate oxidation, and acetylation-dependent mechanisms intersecting drug iatrogenesis. Cell Mol Life Sci 2021;78(23):7451-7468. doi:10.1007/s00018-021-03996-3
- ^ Brautigam C.A., Chuang J.L., Tomchick D.R., Machius M., Chuang D.T. Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. J Mol Biol 2005;350(3):543-52. doi:10.1016/j.jmb.2005.05.014
- ^ a b c Mattevi A., Obmolova G., Sokatch J.R., Betzel C., Hol W.G. The refined crystal structure of Pseudomonas putida lipoamide dehydrogenase complexed with NAD+ at 2.45 A resolution. Proteins 1992;13(4):336-51. doi:10.1002/prot.340130406
- ^ Kern D., Kern G., Neef H., Tittmann K., Killenberg-Jabs M., Wikner C., Schneider G., Hübner G. How thiamine diphosphate is activated in enzymes. Science 1997;275(5296):67-70. doi:10.1126/science.275.5296.67
- ^ a b Jordan F. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat Prod Rep 2003;20(2):184-201. doi:10.1039/b111348h
- ^ a b Nemeria N.S., Chakraborty S., Balakrishnan A., Jordan F. Reaction mechanisms of thiamin diphosphate enzymes: defining states of ionization and tautomerization of the cofactor at individual steps. FEBS J 2009;276(9):2432-46. doi:10.1111/j.1742-4658.2009.06964.x
- ^ Hawkins C.F., Borges A., Perham R.N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett 1989;255(1):77-82. doi:10.1016/0014-5793(89)81064-6
- ^ Breslow R. Rapid deuterium exchange in thiazolium salts. J Am Chem Soc 1957;79:1762-1763. doi:https://doi.org/10.1021/ja01564a064
- ^ Baykal A.T., Kakalis L., Jordan F. Electronic and nuclear magnetic resonance spectroscopic features of the 1′,4′-iminopyrimidine tautomeric form of thiamin diphosphate, a novel intermediate on enzymes requiring this coenzyme. Biochemistry 2006;45(24):7522-8. doi:10.1021/bi060395k
- ^ Meyer D., Neumann P., Koers E., Sjuts H., Lüdtke S., Sheldrick G.M., Ficner R., Tittmann K. Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin. Proc Natl Acad Sci USA 2012;109(27):10867-72. doi:10.1073/pnas.1201280109
- ^ Shaanan B., Chipman D.M. Reaction mechanisms of thiamin diphosphate enzymes: new insights into the role of a conserved glutamate residue. FEBS J 2009 May;276(9):2447-53. doi:10.1111/j.1742-4658.2009.06965.x
- ^ Yang Y.S., Frey P.A. Dihydrolipoyl transacetylase of Escherichia coli. Formation of 8-S-acetyldihydrolipoamide. Biochemistry 1986;25(25):8173-8. doi:10.1021/bi00373a008
- ^ Škerlová J., Berndtsson J., Nolte H., Ott M., Stenmark P. Structure of the native pyruvate dehydrogenase complex reveals the mechanism of substrate insertion. Nat Commun 2021;12(1):5277. doi:10.1038/s41467-021-25570-y
- ^ Massey V., Gibson Q.H., Veeger C. Intermediates in the catalytic action of lipoyl dehydrogenase (diaphorase). Biochem J 1960;77(2):341-51. doi:10.1042/bj0770341
- ^ Rice D.W., Schulz G.E., Guest J.R. Structural relationship between glutathione reductase and lipoamide dehydrogenase. J Mol Biol 1984;174(3):483-96. doi:10.1016/0022-2836(84)90332-2
- ^ Denton R.M., Randle P.J., Bridges B.J., et al. Regulation of mammalian pyruvate dehydrogenase. Mol Cell Biochem 1975;9(1):27-53. doi:10.1007/BF01731731
- ^ Gray L.R., Tompkins S.C., Taylor E.R. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci 2014;71(14):2577-2604. doi:10.1007/s00018-013-1539-2
- ^ Harris R.A., Bowker-Kinley M.M., Huang B., Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Adv Enzyme Regul 2002;42:249-59. doi:10.1016/s0065-2571(01)00061-9
- ^ Kolobova E., Tuganova A., Boulatnikov I., Popov K.M. Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem J 2001;358(Pt 1):69-77. doi:10.1042/0264-6021:3580069
- ^ Hiromasa Y., Fujisawa T., Aso Y., Roche T.E. Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J Biol Chem 2004;279(8):6921-33. doi:10.1074/jbc.M308172200
- ^ Roche T.E., Baker J.C., Yan X., et al. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol 2001;70:33-75. doi:10.1016/s0079-6603(01)70013-x
- ^ a b c Pettit F.H., Pelley J.W., Reed L.J. Regulation of pyruvate dehydrogenase kinase and phosphatase by acetyl-CoA/CoA and NADH/NAD ratios. Biochem Biophys Res Commun 1975;65(2):575-82. doi:10.1016/s0006-291x(75)80185-9
- ^ Patel M.S., Korotchkina L.G. Regulation of the pyruvate dehydrogenase complex. Biochem Soc T 2006;34(2):217-222. doi:10.1042/bst0340217
- ^ Small L., Brandon A.E., Quek L.E., Krycer J.R., James D.E., Turner N., Cooney G.J. Acute activation of pyruvate dehydrogenase increases glucose oxidation in muscle without changing glucose uptake. Am J Physiol Endocrinol Metab 2018;315(2):E258-E266. doi:10.1152/ajpendo.00386.2017