The human gastrointestinal tract is one of the most highly competitive ecological niches in the human body. It harbors a complex ecosystem composed of viruses, eukaryotes, bacteria, and members of the Archaea, including Methanobrevibacter smithii. This ecosystem, collectively referred to as the gut microbiota, represents a major component of the broader human microbiota.[1][2]
The gut microbiota is characterized by a remarkable taxonomic and functional complexity. It comprises hundreds of microbial species organized into structured communities, shaped by ecological interactions among microorganisms, including bacteria, bacteriophages, and other components of the gut ecosystem. Together, these interactions contribute to both community stability and inter-individual variability.[3][4]
Beyond its compositional features, the gut microbiota is a highly dynamic entity whose structure changes profoundly over time. Microbial communities are established early in life, undergo rapid remodeling during infancy and childhood, and progressively evolve through adulthood and aging. Early-life events, particularly the mode of delivery, play a pivotal role in its initial assembly and may influence immune system development and long-term microbial trajectories.[5][6][7]
After this early developmental phase, additional factors progressively shape gut microbiota composition and function. These include spatial differences along the gastrointestinal tract, driven by variations in pH, oxygen availability, digestive enzymes, and bile salts, as well as external modulators such as diet and geographical location. Together, these pressures contribute to the diversification of microbial communities and to their functional specialization.[8][9][10]
As a result, the gut microbiota emerges as a dynamic, spatially organized, and temporally regulated ecosystem. As a central component of the human microbiota, it plays a fundamental role in host physiology and represents a key link between environmental exposures, host biology, and health and disease.[11]
Contents
- Composition of the gut microbiota
- Gut microbiota composition throughout life
- Gut microbiota and intestinal environment
- Gut microbiota and diet
- Gut microbiota, health status, and disease
- Gut microbiota, bacteriocins, and bacteriophages
- Gut microbiota and geographical location
- References
Composition of the gut microbiota
Bacterial abundance and composition vary along the gastrointestinal tract and are particularly high in the colon.
The adult human colon contains more than 400 bacterial species, belonging to nine major phyla out of the approximately 30 currently recognized. However, metagenomic approaches suggest that the actual diversity is substantially higher.[12]
The most abundant bacterial phyla of the Western adult gut microbiota and their representative genera are summarized in the table below.[13][14]
| Phylum | Gram staining | Representative genera |
|---|---|---|
| Actinobacteria | Gram-positive | Bifidobacterium, Collinsella, Corynebacterium, Eggerthella, Propionibacterium |
| Bacteroidetes | Gram-negative | More than 20 genera, including Bacteroides and Prevotella |
| Cyanobacteria | Gram-negative | — |
| Firmicutes | Gram-positive | Mycoplasma, Bacillus, Clostridium, Dorea, Faecalibacterium, Ruminococcus, Eubacterium, Staphylococcus, Streptococcus, Lactobacillus, Lactococcus, Enterococcus, Sporobacter, Roseburia |
| Fusobacteria | Gram-negative | — |
| Lentisphaerae | Gram-negative | — |
| Proteobacteria | Gram-negative | Escherichia, Klebsiella, Shigella, Salmonella, Citrobacter, Helicobacter, Serratia |
| Spirochaetes | Gram-negative | — |
| Verrucomicrobia | Gram-negative | Akkermansia |
Gut ecosystem and the virome
The adult gut harbors a large and diverse community of DNA and RNA viruses, collectively known as the virome, comprising thousands of viral genotypes, none of which is dominant. Indeed, the most abundant virus accounts for only about 6% of the community, whereas in infants the most abundant virus accounts for over 40%.[15][16][17]
The majority of DNA viruses are bacteriophages (or phages), namely viruses that infect bacteria. They are the most abundant biological entities on Earth, with an estimated population of approximately 1031 particles, whereas many RNA viruses detected are plant viruses.[18]
Selective pressure and individual variability
The presence of only a small subset of the bacterial world in the colon is the result of strong selective pressures that have acted during evolution on both microbial colonizers, selecting organisms highly adapted to this environment, and the intestinal niche itself.[3][7] Nevertheless, each individual harbors a unique bacterial community in the gut. This individual specificity coexists with recurrent ecological patterns observed across populations.[19]
Despite the high variability observed both among taxa and between individuals, it has been proposed, although not universally accepted, that in most adults the gut microbiota can be classified into variants, or enterotypes, based on the relative abundance of the genera Bacteroides and Prevotella.[20] This suggests the existence of a limited number of well-balanced symbiotic states, which may respond differently to factors such as intestinal environment, diet, age, genetics, health status, and drug intake.[21]
Gut microbiota composition throughout life
The development of the intestinal microbial ecosystem is a complex and crucial process in human life, and is highly variable among individuals.[6][7]
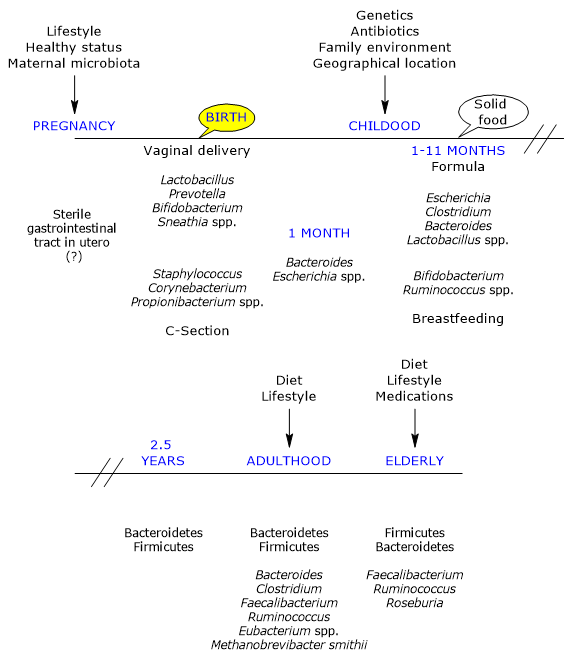
In utero, the gut has traditionally been considered sterile; however, it is rapidly colonized by microbes at birth, as the infant is born with an immunological tolerance shaped by the mother.[22]
Recent studies, however, have reported the presence of bacteria in placental tissue, umbilical cord blood, fetal membranes, and amniotic fluid from healthy newborns without signs of infection or inflammation.[23] For example, the meconium of premature infants born to healthy mothers contains a specific microbiota, with Firmicutes as the main phylum and a predominance of staphylococci. In contrast, Proteobacteria, particularly species such as Escherichia coli, Klebsiella pneumoniae, and Serratia marcescens, as well as enterococci, are more abundant in feces.[24]
It appears that both vaginal and gut bacteria may gain access to the fetus, although through different routes of entry: vaginal bacteria via ascending pathways, and gut bacteria via immune system dendritic cells. Therefore, the existence of a fetal microbiota has been proposed.[6]
Mode of delivery
The first major determinant of postnatal microbial colonization is the mode of delivery.[25]
Colonization occurs during delivery through a maternal inoculum, generally composed of aerobic and facultative bacteria, as the newborn gut initially contains oxygen. These are later replaced by obligate anaerobes, which are typically found in adulthood and for which a suitable environment is gradually established.[26]
The maternal vaginal and fecal microbiotas are the primary sources of inoculum in vaginally delivered infants. Accordingly, infants harbor microbial communities dominated by species of the genera Lactobacillus, the most abundant genus in the vaginal microbiota and early gut microbiota, Bifidobacterium, Prevotella, and Sneathia.[27] Anaerobic bacteria, such as members of the phyla Firmicutes and Bacteroidetes, which cannot grow outside the host, likely rely on close mother–infant contact for transmission. Moreover, due to the presence of oxygen in the infant gut, the transmission of strict anaerobes may not occur directly at birth but later, possibly via spores.[28]
In infants born by caesarean section, the first bacteria encountered originate from the skin and hospital environment. Their gut microbiota is dominated by species of the genera Corynebacterium, Staphylococcus, and Propionibacterium, with a lower bacterial load and diversity during the first weeks of life compared with vaginally delivered infants.[29][30]
Further evidence supporting vertical transmission comes from the similarity between meconium microbiota and samples obtained from potential maternal sources. These maternal bacteria do not persist indefinitely and are replaced by other microbial populations within the first year of life.[31]
Objects, animals, the mouths and skin of relatives, as well as breast milk, serve as secondary sources of inoculum. Among these, breast milk appears to play a primary role in determining microbial succession in the gut. Inter-individual variation among children reflects the uniqueness of these microbial exposures.[32]
Mode of delivery and immune system
Mode of delivery also appears to influence immune system development during the first year of life, possibly through its effects on gut microbiota.[33]
Infants born by caesarean section show:
- a lower bacterial load in stool samples at one month of age, largely due to reduced bifidobacterial abundance;
- a higher number of antibody-secreting cells, potentially reflecting excessive antigen exposure due to increased intestinal permeability.
Together, these findings support the concept that early alterations in microbial colonization associated with caesarean delivery may influence immune maturation during a critical developmental window.[30][34][35]
From birth to first month
Within a few days after birth, a thriving microbial community is established. This community is less stable and more variable than that of adults and soon becomes more numerous than the host’s own cells. Its development follows a highly individual temporal pattern.[5]
During the first month of life, a limited number of taxa, mainly belonging to the phyla Actinobacteria and Proteobacteria, remain relatively stable. In subsequent months, however, there is a marked increase in variability and the emergence of new genetic variants.[6] Numerous studies emphasize that early microbial exposure plays a critical role in shaping the “trajectories” leading to the adult ecosystem. These early communities may also act as sources of either protective or pathogenic microorganisms.[17]
Viruses, absent at birth, reach about 108 particles per gram of wet fecal weight by the end of the first week of life, becoming a dynamic and abundant component of the developing gut ecosystem. Although viral diversity is initially low, the virome is dominated by bacteriophages, which likely influence bacterial abundance and diversity. The origin of these viruses remains unclear, although maternal and environmental sources are plausible.[36] Notably, early viruses may arise from prophage induction within the newborn gut microbiota, as suggested by the observation that over 25% of phage sequences closely resemble those infecting Lactococcus, Lactobacillus, Enterococcus, and Streptococcus, genera abundant in breast milk.[37][38]
From first month to first year
By the end of the first month of life, the initial phase of rapid microbial acquisition is thought to be complete. In 1-month-old infants, the most abundant bacteria belong to the genera Bacteroides and Escherichia. Subsequently, Bifidobacterium, together with Ruminococcus, increases and becomes dominant in the gut of breastfed infants between 1 and 11 months of age.[39][40]
Bifidobacteria, such as Bifidobacterium longum subsp. infantis:
- are closely associated with breastfeeding;
- are among the best-characterized commensal bacteria;
- are considered probiotics, capable of conferring health benefits to the host.[41]
Their abundance also provides protection through competitive exclusion, limiting pathogen colonization. In contrast, Escherichia (e.g., E. coli), Clostridium (e.g., C. difficile), Bacteroides (e.g., B. fragilis), and Lactobacillus are present at higher levels in formula-fed infants than in breastfed infants.[42]
Although breastfed infants receive only breast milk until weaning, their microbiota may exhibit considerable inter-individual variability in both taxonomic abundance and temporal dynamics. These differences may arise from disease, antibiotic exposure, lifestyle changes, stochastic colonization events, and variations in immune responses. However, the relative contribution of each factor remains unclear.[5][43]
The virome also changes rapidly after birth: most viral sequences detected during the first week of life are absent after the second week, and diversity expands markedly during the first three months.[17][44] This contrasts with the adult virome, in which approximately 95% of sequences remain stable over time.[15][45]
From first year to 2.5 years
Under normal conditions, by the end of the first year of life, infants have consumed a diversified, family-type diet for a sufficient period and develop a microbial community resembling that of adults, characterized by:[5]
- increased stability and phylogenetic complexity, with greater similarity among individuals;[46]
- dominance of Firmicutes and Bacteroidetes, followed by Verrucomicrobia and low levels of Proteobacteria;[47]
- increased fecal bacterial load and production of short-chain fatty acids, mainly acetate, propionate, and butyrate;[48]
- enrichment of genes involved in xenobiotic degradation, vitamin biosynthesis, and carbohydrate metabolism.[49]
Despite the substantial taxonomic turnover from birth to one year of age, overall functional capacity remains remarkably constant.[50] By this stage, early viral colonizers are also replaced by a child-specific virome.[44]
The gut microbiota reaches full maturity at approximately 2.5 years of age, closely resembling that of adults. This maturation results from several factors:
- transition to an adult diet;
- greater fitness of adult-associated taxa compared with early colonizers;
- developmental changes in the intestinal mucosa;
- ecological effects exerted by the microbiota itself.
Consequently, the first 2–3 years of life represent a critical window during which targeted interventions may optimize microbiota development and support healthy growth.[7]
Adulthood
From an initially chaotic state, the gut ecosystem evolves into a relatively stable adult configuration, persisting until old age. This ecosystem includes viral, archaeal, and eukaryotic components and, in Western populations, is dominated by Firmicutes (approximately 60%), followed by Bacteroidetes and Actinobacteria, mainly Bifidobacterium, each representing about 10%, with lower proportions of Proteobacteria and Verrucomicrobia. The genera Bacteroides, Clostridium, Faecalibacterium, Ruminococcus, and Eubacterium, together with Methanobrevibacter smithii, constitute the majority of the adult gut microbiota.[51][52]
| Microbial group | Phylum | Representative genera | Approximate abundance |
|---|---|---|---|
| Bacteria | Firmicutes | Clostridium, Faecalibacterium, Ruminococcus, Eubacterium, Roseburia | ~60% |
| Bacteria | Bacteroidetes | Bacteroides, Prevotella | ~20–30% |
| Bacteria | Actinobacteria | Bifidobacterium, Collinsella | ~10% |
| Bacteria | Proteobacteria | Escherichia, Klebsiella, Helicobacter | <5% |
| Bacteria | Verrucomicrobia | Akkermansia | <5% |
| Archaea | Euryarchaeota | Methanobrevibacter | <1% |
| Viruses | — | Bacteriophages | Highly abundant |
Different patterns have been observed in rural African populations.[53] Nevertheless, the gut microbiota is sufficiently conserved across individuals to allow identification of a shared core microbiome.[54]
Stability and resilience are influenced by numerous variables, among which diet appears to be one of the most important. Maintaining microbiota stability therefore requires minimizing disruptive factors and preventing disease, including through vaccination. However, stability may be detrimental if the dominant microbial community is pathogenic.[55][56]
Older age
In elderly individuals, the gut microbiota undergoes significant changes.[57] A study conducted in Ireland on 161 healthy subjects aged 65 years and older showed that, in most individuals, the microbiota differs substantially from that of younger adults. It is dominated by Bacteroidetes and Firmicutes, with nearly inverted proportions compared with younger subjects, although considerable inter-individual variability exists. Faecalibacterium accounts for approximately 6% of dominant genera, followed by Ruminococcus, Roseburia, and Bifidobacterium, the latter representing about 0.4%.[58]
Microbiota variability is also greater in the elderly, possibly due to increased comorbidities, higher medication use, and age-related dietary changes.[51][59]
Gut microbiota and intestinal environment
Due to its low pH, the stomach represents a hostile environment for bacteria, which are present in relatively low numbers, approximately 102–103 bacterial cells per gram of tissue. In addition to Helicobacter pylori, which can cause gastritis and gastric ulcers, microorganisms belonging to the genus Lactobacillus are also detected.[8][60]
Upon reaching the duodenum, bacterial numbers increase to approximately 104−105 cells per gram of tissue, with similar concentrations in the jejunum and proximal ileum.[61] The relatively low microbial load of the small intestine is due to its inhospitable environment, resulting from the opening of the ampulla of Vater in the descending portion of the duodenum, which releases pancreatic juice and bile. Pancreatic enzymes and bile salts exert antimicrobial effects that limit bacterial growth.[10]
In the terminal ileum, where the activity of pancreatic enzymes and bile salts is reduced, bacterial numbers rise to approximately 107 cells per gram of tissue, reaching up to 1012−1014 cells per gram in the colon. As a consequence, bacteria account for a large proportion, about 40%, of fecal mass.[62]
Bacterial distribution along the intestine
The intestinal environment, defined by pH gradients, bile salts, digestive enzymes, oxygen availability, and nutrient distribution, exerts a major selective pressure on the gut microbiota.[63]
The distribution of bacteria along the intestine is strategic. In the duodenum and jejunum, nutrient availability is high, whereas in the terminal ileum and colon only water, fiber, and electrolytes remain. Therefore, high bacterial densities in the distal gut are not problematic.[64] Conversely, elevated bacterial concentrations in the duodenum, jejunum, and proximal ileum are pathological and characterize a condition known as small intestinal bacterial overgrowth (SIBO), in which bacterial numbers increase by one or more orders of magnitude compared with physiological levels. This results in competition with the host for nutrients and leads to gastrointestinal symptoms such as diarrhea.[65]
Gut microbiota and diet
Diet is widely recognized as the most influential factor shaping the composition and metabolic activity of the gut microbiota.[66] Both the quality and the temporal pattern of nutrient intake exert strong selective pressures, affecting microbial diversity from early life through adulthood. Dietary effects can be observed at multiple levels, ranging from initial colonization to long-term enterotype configuration and modulation of the gut virome.[15][55][67]
Role of breast milk
Traditionally considered sterile, human milk harbors a rich microbiota consisting of more than 700 species, dominated by staphylococci, streptococci, bifidobacteria, and lactic acid bacteria.[68] It therefore represents a major source for the colonization of the breastfed infant gut.[32] This mode of colonization has been suggested to be closely correlated with infant health status, as it may protect against infections and contribute to immune system maturation.[69] Breast milk also affects the intestinal microbiota indirectly through the presence of oligosaccharides with prebiotic activity, which stimulate the growth of specific bacterial groups, including bifidobacteria.[70]
Dietary patterns in different populations
A study compared the intestinal microbiota of European and African children, aged between 1 and 6 years, from Florence and a rural village in Burkina Faso, respectively. The study highlighted the dominant role of diet over variables such as climate, geography, hygiene, and health services.[53]
A relative functional convergence has been reported, with no major differences in key immune-related pathways between the two groups. Indeed, as widely reported in the literature, infants, as long as they are breastfed, display a very similar gut microbiota, rich in Actinobacteria, mainly Bifidobacterium.[71]
The subsequent introduction of solid foods, namely, a Western diet rich in animal fats and proteins in European children, and a diet low in animal proteins but rich in complex carbohydrates in African children, leads to a differentiation in the Firmicutes/Bacteroidetes ratio between the two groups. Gram-positive bacteria, mainly Firmicutes, were more abundant in European children, whereas Gram-negative bacteria, mainly Bacteroidetes, prevailed in African children.[72]
Long-term and short-term dietary effects
Long-term dietary patterns are strongly associated with enterotype partitioning. Specifically:
- a diet high in animal fats and proteins (a Western-type diet) leads to a gut microbiota dominated by the Bacteroides enterotype;
- a diet high in complex carbohydrates, typical of agrarian societies, leads to the prevalence of the Prevotella enterotype.[67]
Similar results emerged from the aforementioned study in children. In European subjects, the gut microbiota was enriched in taxa typical of the Bacteroides enterotype, whereas in children from Burkina Faso the Prevotella enterotype prevails.[72]
Short-term dietary changes (approximately 10 days), particularly macronutrient shifts and fiber-driven interventions, result in rapid alterations of gut microbiota composition, detectable within 24 hours. However, these changes do not lead to stable modifications in enterotype partitioning, indicating that long-term dietary patterns are required to induce sustained enterotype transitions.[73]
Diet and virome
Dietary interventions can also induce changes in the gut virome, leading to a new equilibrium state. These changes involve shifts in the relative proportions of pre-existing viral populations, toward which individuals consuming the same diet tend to converge.[15][16]
Gut microbiota, health status, and disease
The health status of both the infant and the mother represents a major determinant of gut microbiota composition.[50] For example, in mothers affected by inflammatory bowel disease (IBD), a condition characterized by chronic intestinal inflammation, the gut microbiota shows a marked depletion of Faecalibacterium prausnitzii, a butyric acid–producing bacterium with well-established anti-inflammatory properties. This reduction is accompanied by an increased abundance of adherent Escherichia coli, a microbial signature commonly associated with intestinal dysbiosis.[74][75]
Gut microbiota, bacteriocins, and bacteriophages
Bacteriocins are proteins with antibacterial activity, whereas bacteriophages play a key role in controlling the abundance and composition of the gut microbiota. In particular, bacteriophages may influence newborn colonization by infecting dominant bacterial populations, thereby allowing other bacterial strains to become prevalent.[76]
This predator–prey dynamic, known as the kill-the-winner model, suggests that blooms of a specific bacterial species are followed by blooms of their corresponding bacteriophages, leading to a subsequent decline in bacterial abundance. Consequently, the most abundant bacteriophage genotype changes over time. Although some gene sequences in the infant gut virome remain stable during the first three months of life, dramatic shifts in the overall viral community composition occur between the first and second week of life. During this period, the bacterial community is also extremely dynamic.[17][37][77]
Gut microbiota and geographical location
Geographical location has been shown to influence gut microbiota composition from early life. In European infants, for example, Northern infants tend to harbor higher levels of Bifidobacterium species and some Clostridium, whereas Southern infants exhibit higher levels of Bacteroides, Lactobacillus, and Eubacterium.[6][78]
More broadly, cross-population studies indicate that geographical location, together with lifestyle factors such as diet, subsistence patterns, and environmental exposures, contributes substantially to variation in gut microbiota composition and diversity across human populations.[79] These findings suggest that both regional environmental influences and culturally driven dietary habits play a key role in shaping the early-life microbiome.
References
- ^ Afzaal M., Saeed F., Shah Y.A., et al. Human gut microbiota in health and disease: unveiling the relationship. Front Microbiol 2022;13:999001. doi:10.3389/fmicb.2022.999001
- ^ Dekaboruah E., Suryavanshi M.V., Chettri D., Verma A.K. Human microbiome: an academic update on human body site specific surveillance and its possible role. Arch Microbiol 2020;202(8):2147-2167. doi:10.1007/s00203-020-01931-x
- ^ a b Ley R.E., Peterson D.A., and Gordon J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006;124(4):837-848. doi:10.1016/j.cell.2006.02.017
- ^ Hitch T.C.A., Hall L.J., Walsh S.K., Leventhal G.E., Slack E., de Wouters T., Walter J., Clavel T. Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol 2022;15(6):1095-1113. doi:10.1038/s41385-022-00564-1
- ^ a b c d Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., and Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol 2007;5(7):e177. doi:10.1371/journal.pbio.0050177
- ^ a b c d e Rodríguez J.M., Murphy K., Stanton C., et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 2015;26:26050. doi:10.3402/mehd.v26.26050
- ^ a b c d Schoultz I., Claesson M.J., Dominguez-Bello M.G., et al. Gut microbiota development across the lifespan: disease links and health-promoting interventions. J Intern Med 2025;297(6):560-583. doi:10.1111/joim.20089
- ^ a b Maurer J.M., Schellekens R.C., van Rieke H.M., et al. Gastrointestinal pH and transit time profiling in healthy volunteers using the IntelliCap system confirms ileo-colonic release of ColoPulse tablets. PLoS One 2015;10(7):e0129076. doi:10.1371/journal.pone.0129076
- ^ Zhang J., Huang Y.J., Yoon J.Y., et al. Primary human colonic mucosal barrier crosstalk with super oxygen-sensitive Faecalibacterium prausnitzii in continuous culture. Med 2021;2(1):74-98.e9. doi:10.1016/j.medj.2020.07.001
- ^ a b Ma H., Wang K., Jiang C. Microbiota-derived bile acid metabolic enzymes and their impacts on host health. Cell Insight 2025;4(5):100265. doi:10.1016/j.cellin.2025.100265
- ^ Zeng Q., Feng X., Hu Y., Su S. The human gut microbiota is associated with host lifestyle: a comprehensive narrative review. Front Microbiol 2025;16:1549160. doi:10.3389/fmicb.2025.1549160
- ^ Qin J., Li R., Raes J., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464(7285):59-65. doi:10.1038/nature08821
- ^ Thursby E., Juge N. Introduction to the human gut microbiota. Biochem J 2017;474(11):1823-1836. doi:10.1042/BCJ20160510
- ^ Almeida A., Nayfach S., Boland M., et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol 2021;39(1):105-114. doi:10.1038/s41587-020-0603-3
- ^ a b c d Minot S., Sinha R., Chen J., Li H., Keilbaugh S.A., Wu G.D., Lewis J.D., and Bushman F.D. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 2011;21:1616-1625. doi:10.1101/gr.122705.111
- ^ a b Cao Z., Sugimura N., Burgermeister E., Ebert M.P., Zuo T., Lan P. The gut virome: a new microbiome component in health and disease. EBioMedicine 2022;81:104113. doi:10.1016/j.ebiom.2022.104113
- ^ a b c d Lim E.S., Zhou Y., Zhao G., Bauer I.K., et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 2015;21(10):1228-34. doi:10.1038/nm.3950
- ^ Batinovic S., Wassef F., Knowler S.A., et al. Bacteriophages in natural and artificial environments. Pathogens 2019;8(3):100. doi:10.3390/pathogens8030100
- ^ Morgan X.C., Segata N., Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet 2013;29(1):51-8. doi:10.1016/j.tig.2012.09.005
- ^ Arumugam M., Raes J., Pelletier E., et al. Enterotypes of the human gut microbiome. Nature 2011;473(7346):174-80. doi:10.1038/nature09944
- ^ Costea P.I., Hildebrand F., Arumugam M., et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol 2018;3(1):8-16. doi:10.1038/s41564-017-0072-8
- ^ Francino M.P. Early development of the gut microbiota and immune health. Pathogens 2014;3(3):769-90. doi:10.3390/pathogens3030769
- ^ Gil A., Rueda R., Ozanne S.E., et al. Is there evidence for bacterial transfer via the placenta and any role in the colonization of the infant gut? – a systematic review. Crit Rev Microbiol 2020;46(5):493-507. doi:10.1080/1040841X.2020.1800587
- ^ Moles L., Gómez M., Heilig H., et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 2013;8(6):e66986. doi:10.1371/journal.pone.0066986
- ^ Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N., and Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci 2010;107:11971-11975. doi:10.1073/pnas.1002601107
- ^ Browne H.P., Neville B.A., Forster S.C., Lawley T.D. Transmission of the gut microbiota: spreading of health. Nat Rev Microbiol. 2017;15(9):531-543. doi:10.1038/nrmicro.2017.50
- ^ Liu T., Kress A.M., Debelius J., et al. Maternal vaginal and fecal microbiota in later pregnancy contribute to child fecal microbiota development in the ECHO cohort. iScience 2025;28(4):112211. doi:10.1016/j.isci.2025.112211
- ^ Rosenberg E., Zilber-Rosenberg I. Reconstitution and transmission of gut microbiomes and their genes between generations. Microorganisms 2021;10(1):70. doi:10.3390/microorganisms10010070
- ^ Goedert J.J. Intestinal microbiota and health of adults who were born by cesarean delivery. JAMA Pediatr 2016;170(10):1027. doi:10.1001/jamapediatrics.2016.2310
- ^ a b Zhang C., Li L., Jin B., Xu X., Zuo X., Li Y., Li Z. The effects of delivery mode on the gut microbiota and health: state of art. Front Microbiol 2021;12:724449. doi:10.3389/fmicb.2021.724449
- ^ Asnicar F., Manara S., Zolfo M., et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2017;2(1):e00164-16. doi:10.1128/mSystems.00164-16
- ^ a b Pannaraj P.S., Li F., Cerini C., et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 2017;171(7):647-654. doi:10.1001/jamapediatrics.2017.0378
- ^ Donald K., Finlay B.B. Early-life interactions between the microbiota and immune system: impact on immune system development and atopic disease. Nat Rev Immunol 2023;23(11):735-748. doi:10.1038/s41577-023-00874-w
- ^ Huurre A., Kalliomäki M., Rautava S., Rinne M., Salminen S., Isolauri E. Mode of delivery-effects on gut microbiota and humoral immunity. Neonatology 2008;93:236-240. doi:10.1159/000111102
- ^ Lai C., Huang L., Wang Y., Huang C., Luo Y., Qin X., Zeng J. Effect of different delivery modes on intestinal microbiota and immune function of neonates. Sci Rep 2024;14(1):17452. doi:10.1038/s41598-024-68599-x
- ^ Breitbart M., Haynes M., Kelley S., et al. Viral diversity and dynamics in an infant gut. Res Microbiol 2008;159:367-373. doi:10.1016/j.resmic.2008.04.006
- ^ a b Mills S., Shanahan F., Stanton C., Hill C., Coffey A., Ross R.P. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 2013;4(1):4-16. doi:10.4161/gmic.22371
- ^ Redgwell T.A., Thorsen J., Petit M.A., et al. Prophages in the infant gut are pervasively induced and may modulate the functionality of their hosts. NPJ Biofilms Microbiomes 2025;11(1):46. doi:10.1038/s41522-025-00674-1
- ^ Koenig J.E., Spor A., Scalfone N., Fricker A.D., Stombaugh J., Knight R., Angenent L.T., and Ley R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci 2011;108(1):4578-4585. doi:10.1073/pnas.1000081107
- ^ Tarracchini C., Milani C., Lugli G.A., Mancabelli L., Turroni F., van Sinderen D., Ventura M. The infant gut microbiota as the cornerstone for future gastrointestinal health. Adv Appl Microbiol 2024;126:93-119. doi:10.1016/bs.aambs.2024.02.001
- ^ Saturio S., Nogacka A.M., Alvarado-Jasso G.M., Salazar N., de Los Reyes-Gavilán C.G., Gueimonde M., Arboleya S. Role of Bifidobacteria on infant health. Microorganisms 2021;9(12):2415. doi:10.3390/microorganisms9122415
- ^ Dargenio V.N., Cristofori F., Brindicci V.F., Schettini F., Dargenio C., Castellaneta S.P., Iannone A., Francavilla R. Impact of Bifidobacterium longum Subspecies infantis on pediatric gut health and nutrition: current evidence and future directions. Nutrients 2024;16(20):3510. doi:10.3390/nu16203510
- ^ Stinson L.F., Norrish I., Mhembere F., Cheema A.S., Mullally C.A., Payne M.S., Geddes D.T. Seeding and feeding: nutrition and birth-associated exposures shape gut microbiome assembly in breastfed infants. Gut Microbes 2025;17(1):2557981. doi:10.1080/19490976.2025.2557981
- ^ a b Kennedy E.A., Holtz L.R. Gut virome in early life: origins and implications. Curr Opin Virol 2022;55:101233. doi:10.1016/j.coviro.2022.101233
- ^ Shkoporov A.N., Clooney A.G., Sutton T.D.S., et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 2019;26(4):527-541.e5. doi:10.1016/j.chom.2019.09.009
- ^ Sekirov I., Russell S.L., Antunes L.C., Finlay B.B. Gut microbiota in health and disease. Physiol Rev 2010;90(3):859-904. doi:10.1152/physrev.00045.2009
- ^ Vallès Y., Artacho A., Pascual-García A., Ferrús M.L., Gosalbes M.J., Abellán J.J., Francino M.P. Microbial succession in the gut: directional trends of taxonomic and functional change in a birth cohort of Spanish infants. PLoS Genet 2014;10(6):e1004406. doi:10.1371/journal.pgen.1004406
- ^ Fusco W., Lorenzo M.B., Cintoni M., et al. Short-chain fatty-acid-producing bacteria: key components of the human gut microbiota. Nutrients 2023;15(9):2211. doi:10.3390/nu15092211
- ^ Derrien M., Alvarez A.S., de Vos W.M. The gut microbiota in the first decade of life. Trends Microbiol 2019;27(12):997-1010. doi:10.1016/j.tim.2019.08.001
- ^ a b Yao Y., Cai X., Ye Y., Wang F., Chen F., Zheng C. The role of microbiota in infant health: from early life to adulthood. Front Immunol 2021;12:708472. doi:10.3389/fimmu.2021.708472
- ^ a b Hou K., Wu Z.X., Chen X.Y., et al. Microbiota in health and diseases. Signal Transduct Target Ther 2022;7(1):135. doi:10.1038/s41392-022-00974-4
- ^ Augustynowicz G., Lasocka M., Szyller H.P., Dziedziak M., Mytych A., Braksator J., Pytrus T. The role of gut microbiota in the development and treatment of obesity and overweight: a literature review. J Clin Med 2025;14(14):4933. doi:10.3390/jcm14144933
- ^ a b De Filippo C., Cavalieri D., Di Paola M., et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci 2010;107(33):14691-14696. doi:10.1073/pnas.1005963107
- ^ Sharon I., Quijada N.M., Pasolli E., et al. The core human microbiome: does it exist and how can we find it? A critical review of the concept. Nutrients 2022;14(14):2872. doi:10.3390/nu14142872
- ^ a b David L.A., Maurice C.F., Carmody R.N., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505(7484):559-63. doi:10.1038/nature12820
- ^ Sommer F., Anderson J.M., Bharti R., Raes J., Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 2017;15(10):630-638. doi:10.1038/nrmicro.2017.58
- ^ Mangiola F., Nicoletti A., Gasbarrini A., Ponziani F.R. Gut microbiota and aging. Eur Rev Med Pharmacol Sci 2018;22(21):7404-7413. doi:10.26355/eurrev_201811_16280
- ^ Claesson M.J., Cusack S., O’Sullivan O., et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA 2011;108(Suppl 1);4586-4591. doi:10.1073/pnas.1000097107
- ^ Salazar N., Valdés-Varela L., González S., Gueimonde M., de Los Reyes-Gavilán C.G. Nutrition and the gut microbiome in the elderly. Gut Microbes. 2017;8(2):82-97. doi:10.1080/19490976.2016.1256525
- ^ Santacroce L., Topi S., Bottalico L., Charitos I.A., Jirillo E. Current knowledge about gastric microbiota with special emphasis on Helicobacter pylori-related gastric conditions. Curr Issues Mol Biol 2024;46(5):4991-5009. doi:10.3390/cimb46050299
- ^ Yang K., Li G., Li Q., et al. Distribution of gut microbiota across intestinal segments and their impact on human physiological and pathological processes. Cell Biosci 2025;15(1):47. doi:10.1186/s13578-025-01385-y
- ^ Portincasa P., Bonfrate L., Khalil M., Angelis M., Calabrese F.M., D’Amato M., Wang D.Q., Di Ciaula A. Intestinal barrier and permeability in health, obesity and NAFLD. Biomedicines 2021;10(1):83. doi:10.3390/biomedicines10010083
- ^ Kastl A.J. Jr, Terry N.A., Wu G.D., Albenberg L.G. The structure and function of the human small intestinal microbiota: current understanding and future directions. Cell Mol Gastroenterol Hepatol 2020;9(1):33-45. doi:10.1016/j.jcmgh.2019.07.006
- ^ Donaldson G.P., Lee S.M., Mazmanian S.K. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 2016;14(1):20-32. doi:10.1038/nrmicro3552
- ^ Leite G., Morales W., Weitsman S., Celly S, et al. The duodenal microbiome is altered in small intestinal bacterial overgrowth. PLoS One 2020;15(7):e0234906. doi:10.1371/journal.pone.0234906
- ^ Zhang L., Tuoliken H., Li J., Gao H. Diet, gut microbiota, and health: a review. Food Sci Biotechnol 2024;34(10):2087-2099. doi:10.1007/s10068-024-01759-x
- ^ a b Wu G.D., Chen J., Hoffmann C., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105-108. doi:10.1126/science.1208344
- ^ Consales A., Cerasani J., Sorrentino G., Morniroli D., Colombo L., Mosca F., Giannì M.L. The hidden universe of human milk microbiome: origin, composition, determinants, role, and future perspectives. Eur J Pediatr 2022;181(5):1811-1820. doi:10.1007/s00431-022-04383-1
- ^ Fernández L., Langa S., Martín V., Maldonado A., Jiménez E., Martín R., Rodríguez J.M. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 2013;69(1):1-10. doi:10.1016/j.phrs.2012.09.001
- ^ Newburg D.S., Morelli L. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr Res 2015;77:115-120. doi:10.1038/pr.2014.178
- ^ Donald K., Finlay B.B. Early-life interactions between the microbiota and immune system: impact on immune system development and atopic disease. Nat Rev Immunol 2023;23(11):735-748. doi:10.1038/s41577-023-00874-w
- ^ a b De Filippo C., Di Paola M., Ramazzotti M., et al. Diet, environments, and gut microbiota. A preliminary investigation in children living in rural and urban Burkina Faso and Italy. Front Microbiol 2017;8:1979. doi:10.3389/fmicb.2017.01979
- ^ Rodriguez C.I., Isobe K., Martiny J.B.H. Short-term dietary fiber interventions produce consistent gut microbiome responses across studies. mSystems 2024;9(6):e0013324. doi:10.1128/msystems.00133-24
- ^ Cao Y., Shen J., Ran Z.H. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Gastroenterol Res Pract 2014;2014:872725. doi:10.1155/2014/872725
- ^ Lavelle A., Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17(4):223-237. doi:10.1038/s41575-019-0258-z
- ^ Emencheta S.C., Olovo C.V., Eze O.C., et al. The role of bacteriophages in the gut microbiota: implications for human health. Pharmaceutics 2023;15(10):2416. doi:10.3390/pharmaceutics15102416
- ^ Mills S., Ross R.P., Hill C. Bacteriocins and bacteriophage; a narrow-minded approach to food and gut microbiology. FEMS Microbiol Rev 2017;41(1):S129-S153. doi:10.1093/femsre/fux022
- ^ Fallani M., Young D., Scott J., et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr 2010;51(1):77-84. doi:10.1097/MPG.0b013e3181d1b11e
- ^ Gupta V.K., Paul S., Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol 2017;8:1162. doi:10.3389/fmicb.2017.01162